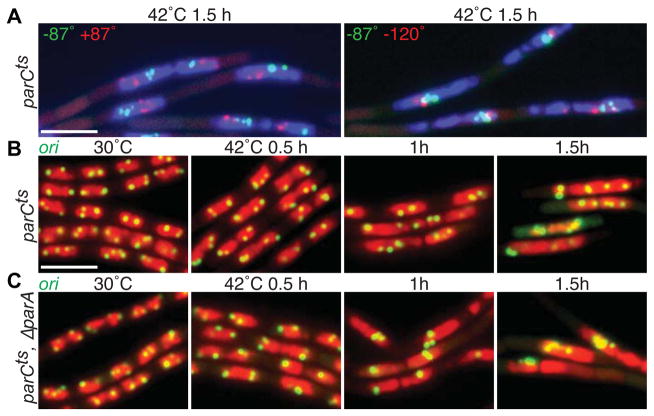
Figure 3. Topoisomerase IV is essential for bulk chromosome segregation but is not necessary to resolve replicated origins.
(A) Representative images of DAPI-stained nucleoids (blue) and indicated chromosomal loci (green and red) in a parCts mutant (BWX2112 and BWX2106) grown in CH medium for 1.5 h after shifting to 42°C. Clusters of green and red foci reflect a failure to segregate the replicated chromosomes. 70–75% (n>1190 per strain) had nucleoids and foci similar to those shown in (A). (B) Representative images of nucleoids (red) and origin loci (green) in a parCts mutant (BWX2082) grown at 30°C and after shift to 42°C. (C) The same time course as in (B) with a parCts mutant that also lacks ParA (BWX2574). Origin foci are more often clustered in the absence of ParA although this is not as pronounced as upon SMC inactivation (Figure 1). 70% (n=1143) of parCts and 19% (n=1238) of ΔparA, parCts had nucleoids with 3 or more well-resolved foci or clusters of foci at hour 1.5. Scale bars are 4 μm.