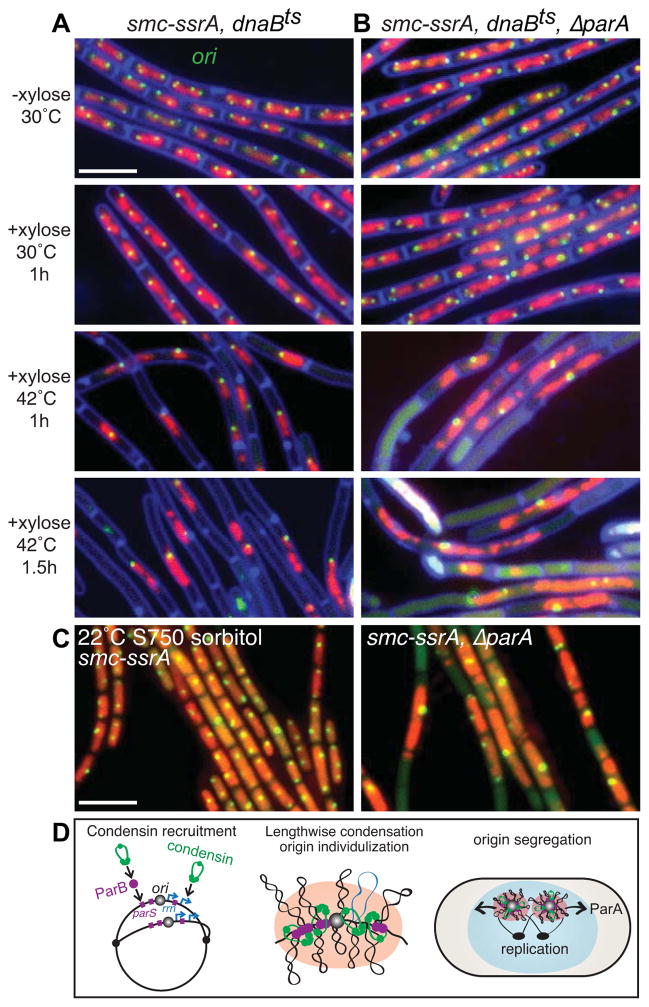
Figure 4. The parABS partitioning system and the SMC complex contribute to origin segregation.
(A) Chromosome resolution and segregation occur in the absence of SMC if new rounds of replication are blocked. Representative micrographs of DAPI-stained nucleoids (false-colored red) and origin foci (green) in a strain (BWX1527) harboring the smc-ssrA degradable allele and a temperature-sensitive replication initiation mutant (dnaBts). After induction of SMC-SsrA degradation for 1 h most cells had unsegregated nucleoids with unresolved origin foci. Inhibition of replication leads to resolution and segregation of the chromosomes if the parABS system is intact (61%; n=1284) (A) but not if ParA is absent (strain BWX2558) (29%; n=1262) (B). (C) Cells lacking SMC and ParA grown under permissive conditions have severe defects in origin segregation. Cells with intact parABS (BWX1497) and lacking parA (BWX2551) were induced to degraded SMC-SsrA under permissive growth conditions (minimal S750 medium supplemented with sorbitol at 22°C). Representative images of nucleoids (false-colored red) and origin foci (green) are shown. 11% (n=1595) of smc-ssrA cells and 51% (n=1287) of ΔparA, smc-ssrA cells had unsegregated nucleoids. 2% and 30%, respectively had a single bright origin focus. Scale bars are 4 μm. (D) Schematic model depicting origin segregation in B. subtilis. The SMC condensin complex (green) is recruited to the origin by ParB (purple) bound to origin-proximal parS sites and is enriched at highly transcribed genes including ribosomal RNA genes (rrn) (blue) that reside adjacent to the origin [10, 11]. Compaction of contiguous DNA segments leads to individualization of the sister origins that are resolved and segregated, in part, through the action of ParA acting on ParB/parS complexes. See also Figure S3.