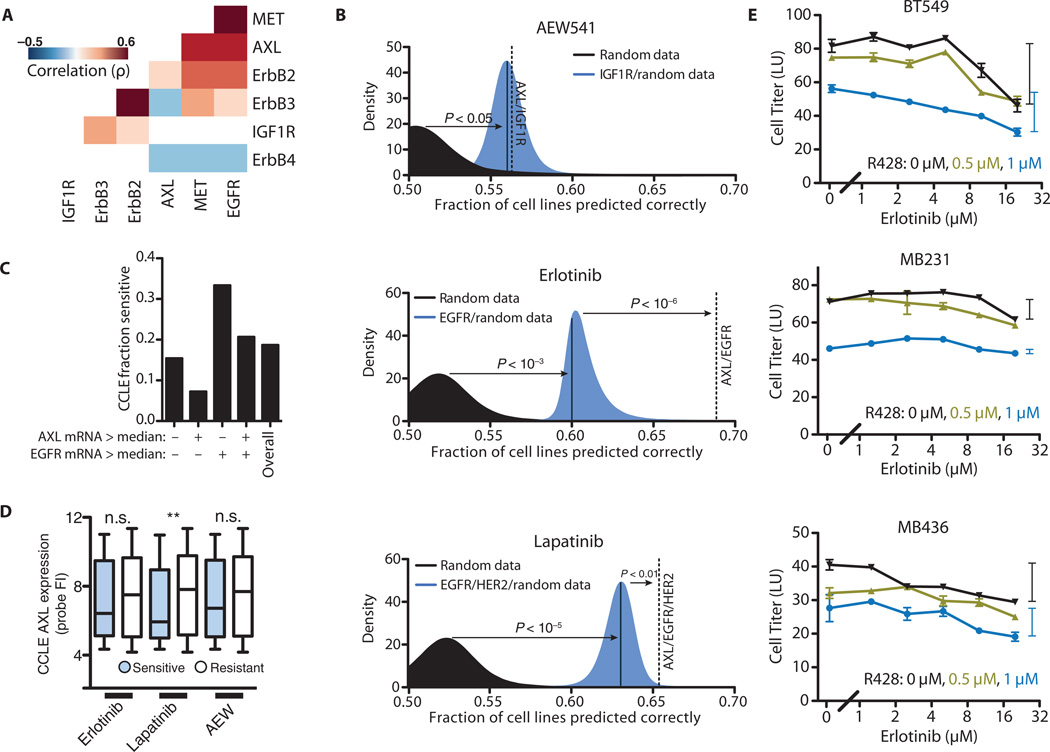
Fig. 1. Support vector classification to identify mechanisms of drug resistance.
(A) Spearman correlations of expression for a subset of RTKs. Only statistically significant correlations are shown (P < 0.01). (B) Classification of cell lines as resistant or sensitive to AEW541, erlotinib, and lapatinib based on RTK expression. Classification accuracy using randomized expression data (black), a model considering the expression of the gene encoding the drug target receptor (blue), or a model considering the expression of both the gene encoding the drug target receptor and that of AXL (dotted line) are shown. (C) Fraction of cell lines that are sensitive to erlotinib after separation according to those that exhibit greater or less than median expression of EGFR or AXL. (D) AXL expression probe values for resistant and sensitive cell lines to each drug (**P < 0.01, Kruskal-Wallis test, n = 91 to 396 cell lines per grouping). FI, fluorescence intensity; n.s., not significant. (E) Dose-response curves for R428 and erlotinib in three TNBC cell lines that have abundant EGFR and AXL. Bars on the side indicate the range of viability between the highest and lowest erlotinib dose to illustrate subadditivity (P < 10−6, BT549; P < 0.05, MB436; P < 0.01, MB231 by Loewe’s synergy analysis; see Materials and Methods) between erlotinib and R428. Data are means ± SEM from three independent biological measurements.