Figure 1.
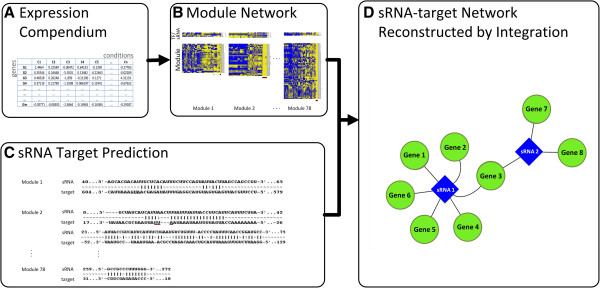
Reconstructing the combined transcriptional-sRNA network. An expression compendium compiled from publicly available microarray data is used as input (showed in Panel A). Using this compendium coexpression modules were constructed by means of biclustering. To each of those modules a regulatory program was assigned using either LeMoNe or CLR (Panel B). For genes in the modules we tested whether they contained a region in their sequence that shows complementarity to any of the sRNA sequences assigned to the module (Panel C). Integration of both the module network (modules with their regulatory programs) and the sequence-based predictions results in a final sRNA-target network (Panel D).
