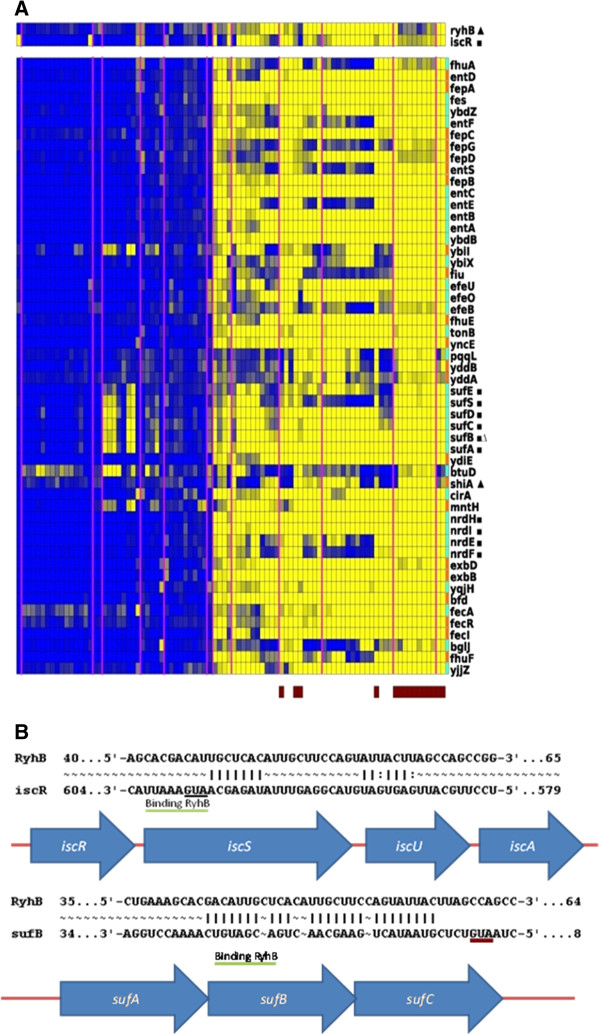
Figure 4.
Module 6 and its regulatory program. Panel A: the regulatory program assigned to this module. The module to which the regulatory program was assigned; yellow indicates high expression levels and blue refers to low expression levels of genes in the module. The genes correspond to the genes present in the original module discovered by ISA. Conditions present in the original ISA module are indicated by a horizontal bar. As both LeMoNe and CLR use all conditions when assigning their respective regulatory program, we indicated also the additional compendium conditions that were relevant for assigning the respective regulatory programs. Genes in the module correspond to likely targets of the assigned regulators. Targets indicated by a square correspond to known targets of the assigned TF (s). Targets indicated by empty triangle correspond to predicted targets of the assigned sRNA, targets indicated by filled triangle correspond to known targets of the assigned sRNA. Panel B: sRNA-target interaction as predicted by the sequence-based analysis for both known and predicted targets of the sRNAs assigned to the module. Indicated sequence positions refer to the location of the recognition sequence relative to the translation start of the gene the sRNA is predicted to interact with (if ATG is indicated in red). If the ATG is indicated in black it refers to the start codon of a neighboring gene. In this case the underlined ATG corresponds to the start codon of IscS as the target site of RyhB is located in the intergenic region between the predicted target IscR and IscS.