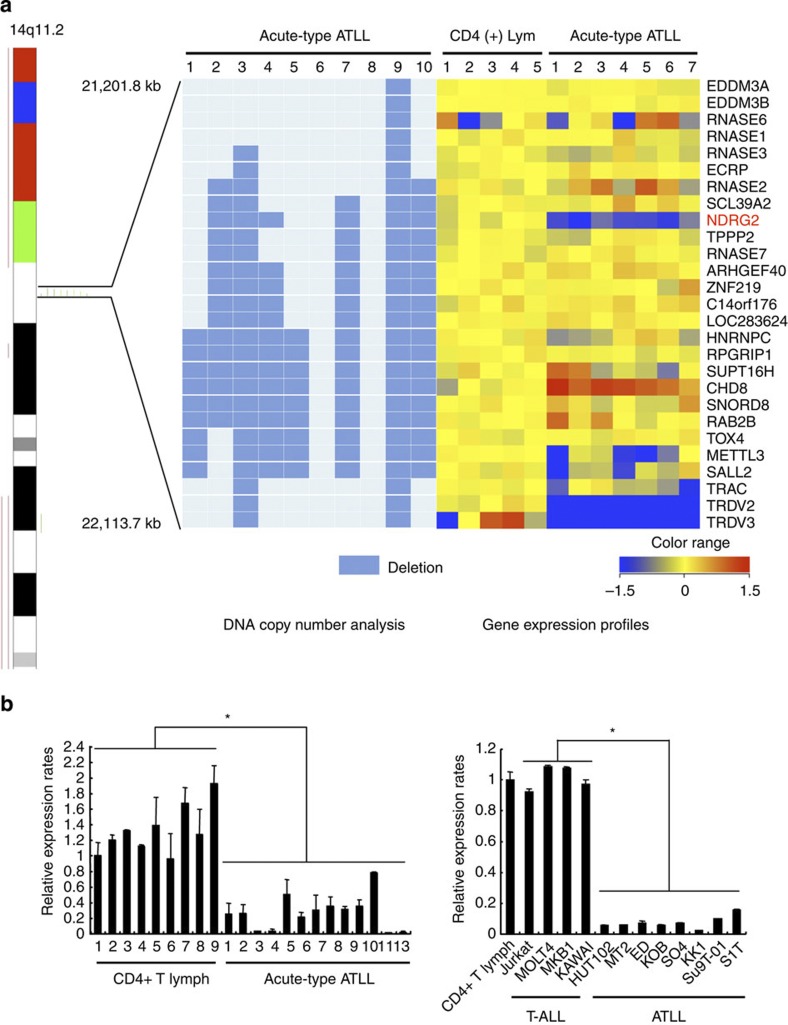
Figure 1. NDRG2 is a candidate tumour suppressor gene in ATLL.
(a) Genomic and gene expression analysis of chromosome 14q11. Results from the SNP array–based comparative genomic hybridization (CGH) analysis on chromosome 14 in ten samples from acute-type ATLL patients. Green bars represent loss of copy number and red bars represent gain of copy number. Out of the ten ATLL samples, eight had breakpoints on 14q11 clustered in a region of 0.9 Mb between genomic positions 21,201,800 and 22,113,700. A heatmap of the normalized gene expression measures for the 27 genes mapped to the recurrent breakpoint region in the CD4+ T lymphocytes from five healthy volunteers and ATLL cells from seven acute-type ATLL patients is shown with the deletion map, in which columns represent samples and rows represent genes. A gradient of blue and red colours represent low- and high-relative fold changes of gene expression to the average expression in normal controls. Genes with average signal intensities less than 100 were eliminated. (b) Expression of NDRG2 mRNA in ATLL cells. Quantitative RT–PCR analysis of NDRG2 was performed with mRNA isolated from nine samples of CD4+ T lymphocytes from healthy volunteers and 13 samples of ATLL cells from the patients, along with samples from four T-ALL cell lines and eight ATLL cell lines. The relative amounts of mRNA were normalized against β-actin mRNA and expressed relative to the mRNA abundance in healthy control sample 1. Mean±s.d. is shown, *P<0.05 (Mann–Whitney U-test). More than a 50% reduction in NDRG2 mRNA expression was observed in 12 out of 13 cases (92%) of ATLL. Data are representative of two experiments.