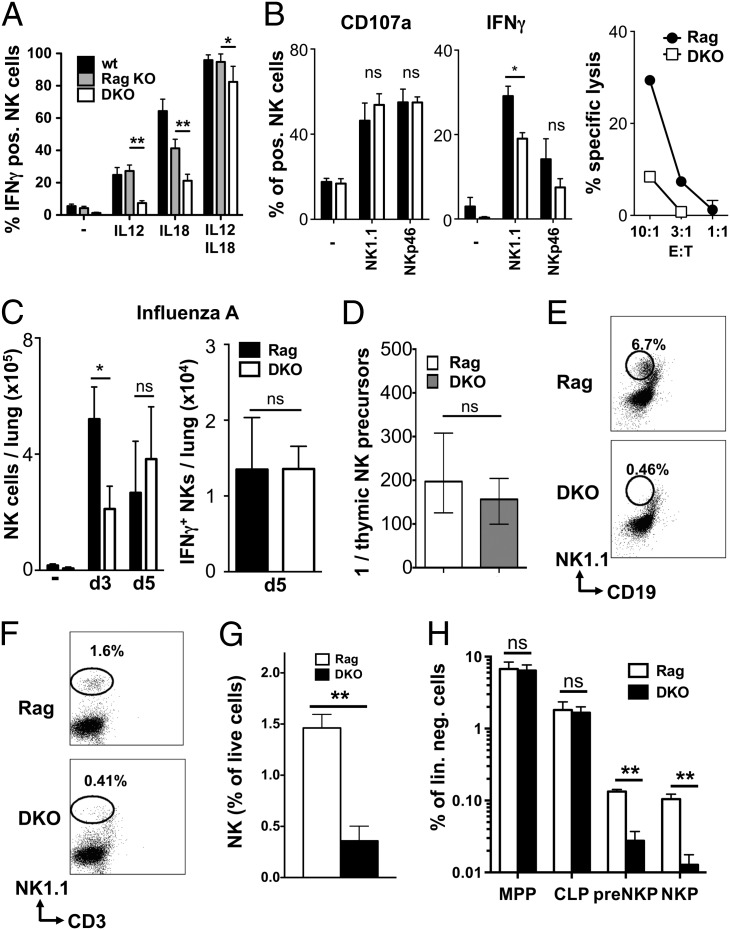
FIGURE 5.
Functionality and origin of NK cells in E4BP4/Rag-1 DKO mice. (A) Total live splenocytes from wt, Rag-1–deficient, or DKO mice were exposed for 16 h to the indicated cytokines in the presence of brefeldin A, and IFN-γ production by NK cells was measured by intracellular cytokine staining (n = 3). Data shown are representative of three similar experiments. (B) Splenic wt and E4BP4−/− NK cells were activated for 12 h with plate-bound anti-NK1.1 or anti-NKp46 Abs and evaluated for IFN-γ production and degranulation (as determined by CD107a expression). Results are representative of three independent experiments. For cytotoxicity, NK cells were mixed at the indicated ratio with YAC-1 target cells, and specific lysis was determined by net lactate dehydrogenase release from target cells. (C) Lungs from uninfected (−), 3-d, or 5-d influenza-infected mice of indicated genotypes were prepared for enumeration of total and IFN-γ+ NK cells by flow cytometry. (D) Determination of the frequency of NK precursors in total lin−CD3−CD4−CD8− DN1 thymocytes by limiting dilution analysis. Sorted cells were plated at frequencies varying from 30 to 3000 cells/well, and numbers of NK cell–positive and –negative wells were used to calculate precursor frequencies. Data from three experiments were pooled for this analysis. (E) Lineage-depleted BM from the indicated mice was cultured first for 5 d in SCF, Flt3L, and IL-7 and then for 7 d in IL-15 with OP9 stromal cells. Cells were then harvested and examined by flow cytometry, gating by scatter and on OP9-GFP–negative cells. The frequency of NK1.1+CD19− NK cells among live GFP− cells is shown. Result representative of three independent experiments. (F and G) Frequency of mature CD3−NK1.1+ NK cells in the BM of DKO or Rag-1–deficient control mice (Rag). Individual plots (F) and mean of three mice (G) are shown. (H) Flow cytometric quantification of frequencies of LMPP (lin−Sca1+ckit+Flt3+CD127−), CLP (lin−Sca1intckitintFlt3+CD127+), pre-NKP (lin−CD27+2B4+CD127+Flt3−CD122−), and refined NKP (lin−CD27+2B4+CD127+Flt3−CD122+) in BM from the indicated mice. Means of three mice are depicted. Error bars indicate SD of replicates (unpaired t test: *p < 0.05, **p < 0.01). ns, not significant.