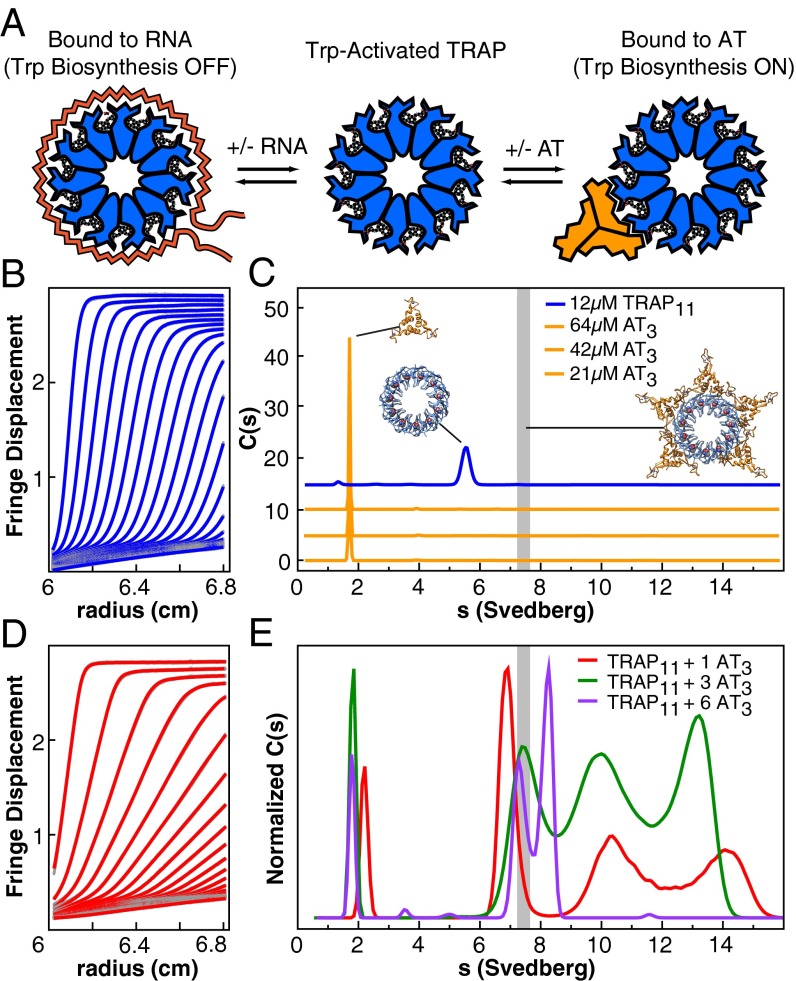
Fig. 1.
In B. subtilis the oligomeric proteins TRAP and AT integratively regulate gene expression through interactions with RNA, Trp, and each other; analytical ultracentrifugation helps describe these interactions. (A) Trp binding activates TRAP for binding to RNA, leading to transcriptional attenuation and translational repression. AT competes with RNA for Trp-activated TRAP, thereby relieving repression. (B) Fringe displacement traces from sedimentation velocity experiments of TRAP alone, illustrating a narrow boundary and homogeneous sedimentation. (C) Continuous sedimentation coefficient distributions for TRAP and AT. A range of concentrations is dominated by single species for each component. Cartoons reflect the AT3 NMR structure, the crystal structure of TRAP11, and a model of 5AT3 bound to TRAP11. The vertical gray bar represents the predicted sedimentation coefficient for the putative TRAP11–5AT3 complex. (D) Fringe displacement traces recorded at an AT3:TRAP11 ratio of 1 reveal the presence of heterogeneous, rapidly sedimenting species. (E) Sedimentation coefficient distributions for samples assembled at AT3:TRAP11 ratios of 1, 3, and 6. Multiple species and/or reaction boundaries are observed. Experiments were performed at pH 7; sedimentation experiments at pH 8 are shown in Fig. S3.