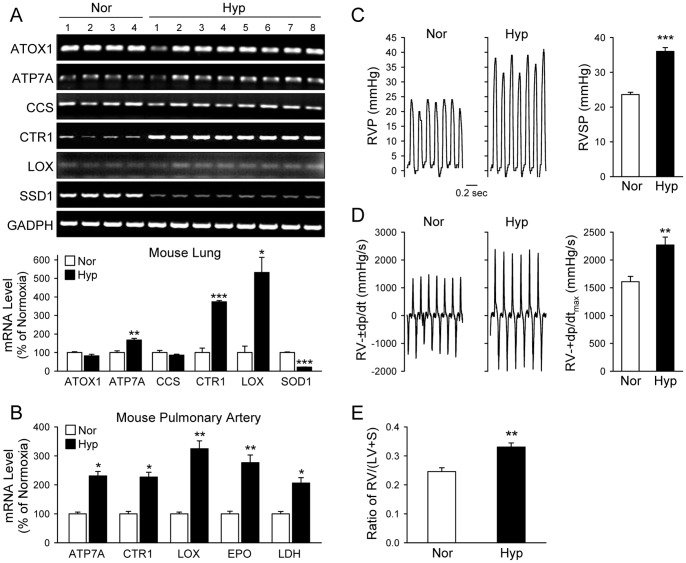
Figure 1. The mRNA expression level of Cu transporters (CTR1 and ATP7A) and lysyl oxidase (LOX) is increased in whole-lung and pulmonary artery (PA) tissues of mice with chronically hypoxia-induced pulmonary hypertension (HPH).
Whole lung tissues and isolated PA tissue from normoxic (Nor, 21% O2) and hypoxic (Hyp, 10% O2 for 5 weeks) mice were homogenized and their mRNA transcripts evaluated by RT-PCR utilizing primers specific for ATOX1, ATP7A, CCS, CTR1, LOX, GAPDH or 18s rRNA (internal controls). A: RT-PCR products from whole-lung tissues were separated on 2% agarose gels (upper panel) and the band intensities quantitated by ImageJ, normalized to intensity of GAPDH, and graphed relative to Nor (n = 4 Nor mouse lungs; n = 8 Hyp mouse lungs). B: PA dissected from Nor and Hyp mice were used for RNA extraction (n = 5) and analyzed by quantitative PCR. Real-time PCR reaction was set with primers specific for the indicated genes. The cycle threshold C(t) values were normalized to 18s rRNA to obtain ΔC(t), quantified relative to normoxic control for each of the indicated genes (ΔΔC(t)), and graphed as % of normoxic control. C: Representative records of right ventricular pressure (RVP, left panel) and summarized data (mean±SE) showing RV systolic pressure (RVSP) in Nor (n = 6) and Hyp (n = 13) mice. D: Representative records (left panel) and summarized data (right panel, mean±SE) of right ventricular contractility (RV-±dp/dtmax) in Nor and Hyp mice. E: Summarized data (mean±SE) showing the ratio of right ventricle (RV) weight to left ventricle (LV) and septum (S) weight [RV/(LV+S)] in Nor (n = 7) and Hyp (n = 7) mice. *P<0.05, **P<0.01, ***P<0.001 vs. Nor.