Abstract
Steroid hormones trigger a wide variety of biological responses through stage- and tissue-specific activation of target gene expression. The mechanisms that provide specificity to systemically released pulses of steroids, however, remain poorly understood. We previously completed a forward genetic screen for mutations that disrupt the destruction of larval salivary glands during metamorphosis in Drosophila melanogaster, a process triggered by the steroid hormone 20-hydroxyecdysone (ecdysone). Here, we characterize 10 complementation groups mapped to genes from this screen. Most of these mutations disrupt the ecdysone-induced expression of death activators, thereby failing to initiate tissue destruction. However, other responses to ecdysone, even within salivary glands, occur normally in mutant animals. Many of these newly identified regulators of ecdysone signaling, including brwd3, med12, med24, pak, and psg2, represent novel components of the ecdysone-triggered transcriptional hierarchy. These genes function combinatorially to provide specificity to ecdysone pulses, amplifying the hormonal cue in a stage-, tissue-, and target gene-specific manner. Most of the ecdysone response genes identified in this screen encode homologs of mammalian nuclear receptor coregulators, demonstrating an unexpected degree of functional conservation in the mechanisms that regulate steroid signaling between insects and mammals.
Keywords: ecdysone, specificity, transcription, cell death, salivary glands
STEROID hormones regulate developmental and homeostatic processes through their respective nuclear receptors, directly regulating expression of critical biological effector genes. Nuclear receptor activity is, in turn, modulated by specific coregulators (Lonard and O’Malley 2007). These coregulators may interact directly or indirectly with nuclear receptors and can increase (coactivators) or decrease (corepressors) the rate of target gene transcription. Importantly, nuclear receptor coregulators have been proposed to act in a combinatorial manner to provide specificity to hormone-induced gene expression (Mckenna and O’Malley 2002; Rosenfeld 2006; Lonard and O’Malley 2007), helping refine global hormonal cues into distinct local responses. However, our understanding of nuclear receptor coregulator function has been primarily restricted to biochemical and tissue culture studies in mammalian systems, making it difficult to define their role in regulating specificity to hormonal cues within the context of a developing animal.
In Drosophila melanogaster, pulses of the steroid hormone 20-hydroxyecdysone (ecdysone) regulate the major developmental transitions during its life cycle. Each pulse triggers a wide variety of biological responses that end one developmental stage and start the next. Like steroid hormones in mammals, ecdysone acts by binding to its nuclear receptor, the ecdysone receptor (EcR), thereby directly regulating target gene expression. Transcriptional targets of ecdysone have been historically divided into two groups: primary response genes, whose expression starts within minutes of hormone exposure and does not require protein synthesis, and secondary response genes, whose expression starts a couple of hours later and requires protein synthesis (Thummel 1996). This hierarchical model of steroid-triggered transcriptional responses was originally proposed by Michael Ashburner and colleagues to explain the behavior of polytene chromosome puffs in insect larval salivary glands (Ashburner et al. 1974). Thus, according to this model, ecdysone initiates transcription of a small number of primary response genes that, in turn, encode transcription factors that initiate expression of a larger set of secondary response genes, many of which were postulated to include regulators of biological processes. However, the relationship, if any, between this hierarchical model and the combinatorial model used to describe mammalian steroid responses has been unclear. These two conceptual frameworks for steroid action have been further confounded by the lack of evidence that nuclear receptor coregulators play a critical role in refining hormonal cues in invertebrates like Drosophila.
Our studies focus on a series of coordinated biological responses to the first two pulses of ecdysone during Drosophila metamorphosis. The late larval pulse of ecdysone triggers puparium formation, ending larval development and initiating prepupal development; ∼12 hr later, the prepupal pulse of ecdysone triggers head eversion, marking the end of prepupal development and the beginning of pupal development (Riddiford 1993). This latter pulse of ecdysone also triggers destruction of larval salivary glands by initiating the stage- and tissue-specific activation of autophagy and caspases (Jiang et al. 1997, 2000; Lee and Baehrecke 2001). This death response depends on the EcR and three ecdysone primary response genes: E74A, br-Z1, and E93 (Jiang et al. 2000; Lee et al. 2000, 2002b). E74A and br-Z1 are induced after both the larval and prepupal pulses of ecdysone (Thummel et al. 1990; Karim and Thummel 1992) and thus are unlikely by themselves to provide the needed specificity to the death response. E93, on the other hand, is induced only after the prepupal pulse and may provide stage specificity to ecdysone even though this expression is not limited to the salivary glands (Lee et al. 2000). Importantly, these events at the onset of metamorphosis provide an ideal context within which to study the mechanisms that provide specificity to ecdysone-triggered biological responses.
To dissect the genetic control of specificity to steroid-triggered responses, we previously conducted a large-scale EMS-mutagenesis screen for mutations that disrupt the destruction of larval salivary glands (Wang et al. 2008). We generated >8600 lethal mutations and identified among them 37 complementation groups that had a persistent salivary gland (PSG) phenotype in otherwise normal-looking animals (Wang et al. 2008). Here, we characterize 10 complementation groups mapped to genes from this screen. Our results demonstrate that these mutations disrupt the death response in salivary glands without disrupting many other ecdysone-triggered responses during metamorphosis. For most PSG mutations, this selectivity is achieved by interrupting ecdysone signaling during the death response, with the mutations mapping to known and novel components of the ecdysone-triggered transcriptional hierarchy. We demonstrate that brwd3, med12, med24, pak, and psg2 encode novel ecdysone secondary response genes required for robust expression of biological effector genes that mediate the death response. Our results indicate that these secondary response genes define an additional regulatory layer within the ecdysone-triggered transcriptional cascade that resides between primary response genes and biological effector genes. Importantly, most of these newly identified ecdysone secondary response genes encode homologs of vertebrate nuclear receptor coregulators, demonstrating a functional relationship between the hierarchical and combinatorial models of steroid action.
Materials and Methods
Drosophila strains
The Bloomington Drosophila Stock Center (BDSC) at Indiana University provided the following stocks: Df(3R)Exel6149, Df(3R)BSC638, Df(3R)Exel6199, BRWD305842, Df(3R)BSC677, Df(3R)Exel6188, Df(3R)BSC523, Eip93Fvno, Df(3R)BSC503, Df(3R)ED6332, hdcBG00237, hdcMI00406, hdcEY02460, Df(3R)Exel6178, Df(3L)ED229, Df(3L)ED228, Df(3L)BSC445, kto1, Df(3L)Exel6112, Df(3R)BSC681, Df(3R)BSC745, Df(3R)BSC744, pak6, pak14, pak16, Df(3L)Exel6105, Df(3L)fz2, Df(3L)ED4789, Df(3L)ED4789, reptEY12756, reptf01801, rept06945, E74Aneo24, Df(3L)81k19, brrbp5, hs-GAL4, UAS-GAL4, Sgs3-GAL4, UAS-EcRF645A, and UAS-p35. Also from the BDSC but not listed are the complementing and lethal mutations used for mapping purposes. K. Moses (Janelia Farm, Asburn, VA) kindly provided the brwd3/ramshackle stocks ram1, ramp1, ramp2, and rampx3. C. Thummel (University of Utah, Salt Lake City, UT) kindly provided the hs-E74A stock. E. Baehrecke (UMass Medical School, Worcester MA) kindly provided the E931 and E936 stocks. The UAS-Atg1K38Q stock was generated in the Nuefeld laboratory (Scott et al. 2007). All PSG mutations were generated in an EMS-mutagenesis screen on the third chromosome (Wang et al. 2008). The allelic combinations used here for phenotypic and molecular analysis of each PSG locus are underlined in Table 1.
Table 1. Summary of PSG complementation groups mapped to genes.
| Failed to complement | |||||||
|---|---|---|---|---|---|---|---|
| Gene | PSG loci | Allele(s) | Cytological position | Deficiencies | Mutations | Sequenced lesions | Source or reference |
| belle | psg9 | psg9 | 85A | Df(3R)p712 | bel6 | psg9: F469S | Ihry et al. (2012) |
| Df(3R)Exel6149 | belL4740 | ||||||
| Df(3R)BSC197 | |||||||
| brwd3 | psg13 | psg13 | 95F | Df(3R)BSC638 | BRWD305842 | psg13: 1222(+4X)a | This study |
| Df(3R)Exel6199 | ram1, ramp1 | ||||||
| ramp2, rampx3 | |||||||
| Eip93F | psg11 | psg11 | 93F | Df(3R)BSC677 | Eip93F6 | psg11: Q485X | This study |
| Df(3R)Exel6188 | Eip93FVno | ||||||
| Df(3R)BSC523 | |||||||
| hdc | psg12 | psg12 | 99F | Df(3R)BSC503 | hdcBG00237 | psg12: Q415X | This study |
| Df(3R)ED6332 | hdcMI00406 | ||||||
| hdcEY02460 | |||||||
| mdh2 | psg7 | psg7ab | 90F | Df(3R)Exel6178 | mdh2EY01940 | psg7a: D173G | Wang et al. (2008) |
| psg7bb | Df(3R)LK19-1 | psg7b: Δ243-253 | |||||
| psg7c | psg7c: N117I | ||||||
| med12 | psg14 | psg14 | 76D | Df(3L)ED229 | kto1 | psg14: Q2175X | This study |
| Df(3L)ED228 | |||||||
| Df(3L)BSC445 | |||||||
| med24 | psg5 | psg5ab | 66B | Df(3L)ED4408 | med24BG01670 | psg5a: M532I, Q893X | Wang et al. (2008) |
| psg5bb | Df(3L)Exel6112 | psg5b: Q212X | |||||
| Df(3L)66C-G28 | |||||||
| pak | psg4 | psg4a | 83E | Df(3R)BSC681 | pak6 | psg4a: Q325X | This study |
| psg4b | Df(3R)BSC745 | pak14 | psg4b: G439S | ||||
| psg4c | Df(3R)BSC744 | pak16 | psg4c: Δ512-604 | ||||
| psg2 | psg2 | psg2a | 64D | Df(3L)Exel6105 | psg2EY04451 | psg2a: Q567X | Wang et al. (2008) |
| psg2b | Df(3L)ZN47 | psg2b: Q650X | |||||
| reptin | psg10 | psg10 | 76A | Df(3L)fz2 | reptEY12756 | psg10: I7V, P318L, V469M | This study |
| Df(3L)ED4789 | reptf01801 | ||||||
| rept06945 | |||||||
Underlined alleles and deficiencies indicate the allelic combinations used for phenotypic and molecular analysis.
Donor splice site mutation in brwd3psg13 that causes a premature stop codon after translating four ectopic residues within the unspliced intron.
PSG alleles previously reported under different names: med241, med24psg5b; mdh22, mdh2psg7a; mdh21, mdh2psg7b.
Mapping PSG mutations
Recombination mapping was conducted using pairs of dominant markers (Sapiro et al. 2013). The resulting recombination position was used to cross each mutation to deficiencies within the DK3 kit in the BDSC and, once mapped to a large deficiency, to progressively smaller deficiencies until a molecularly defined region was identified for each complementation group. Then, all publicly available lethal mutations in the BDSC within these defined regions were tested for complementation. To verify the gene assignment, we tested a number of alleles of the gene in question for complementation and used Sanger sequencing to identify lesions within genomic DNA. The mapping data are summarized in Table 1.
Developmental staging and PSG phenotypes
Animals were reared on cornmeal molasses media with yeast at 25°. Mutant animals were collected 7–10 days after seeding of bottles containing fresh media. For staging during metamorphosis, animals were synchronized as white prepupae [0 hr after puparium formation (0 APF)] and aged appropriately on moist black filter paper on petri dishes at 25°. Average duration of prepupal development was used to determine the staging prior to head eversion. For staging during late larval stages, animals were reared in media containing bromophenol blue to select dark gut (−12 APF), light gut (−8 APF), and clear gut (−4 APF) stages (Andres and Thummel 1994). Block in the destruction of salivary glands was assayed at +24 APF in mutant animals carrying the fkh-GAL4, UAS-GFP transgenes as previously described (Ward et al. 2003). Only persistent salivary glands in animals that head everted relatively normally were counted as having a defect in the larval salivary gland death response. Lethal phases were determined using Bainbridge and Bownes stages (Bainbridge and Bownes 1981): P1–P3 for prepupae (PP), P4–P9 for pupae (P), P10–P15 for pharate adults (PA), and A1–A3 for eclosed adults (A). Images of dissected pupae and of persistent salivary glands were captured using an Olympus SZX16 stereomicroscope coupled to a digital camera. Staging and heat-shock treatments for ectopic expression of E74A were performed with control and mutant animals carrying one copy of the hs-E74A transgene as previously described (Ihry et al. 2012).
Quantitative RT-PCR
To quantify messenger RNA (mRNA) of target genes in whole animals and dissected salivary glands, quantitative real-time reverse transcriptase PCR (qPCR) was performed as previously described (Ihry et al. 2012). RNA was isolated using the RNeasy Plus Mini kit (QIAGEN, Valencia, CA). For each cDNA synthesis reaction, the SuperScript III First-Strand Synthesis System was used with 400 ng of total RNA template primed with Oligo(dT)20 (Invitrogen, Carlsbad, CA). qPCR was conducted using the Roche 480 LightCycler and LightCycler 480 DNA SYBR Green I Master kit. In all cases, experimental samples and matched controls were analyzed simultaneously for expression of target genes and the reference gene rp49. Primer sequences for reaper, hid, Nc, Ark, E74A, E75A, E93, and rp49 were previously described (Ihry et al. 2012); the remaining primers used in this study are listed in Supporting Information, Table S1. Relative expression was calculated using the Relative Expression Software Tool (REST) (Pfaffl et al. 2002) as previously described (Ihry et al. 2012). Each data point used represents the analysis of three independent biological samples (except as indicated in Figure 1B). For developmental expression profiles, the relative expression for each target gene was compared to samples from the stage with the lowest expression. The expression profiles and bar graphs in Figure 6 were compared to values for control salivary glands at 4 hr before head eversion [-4 hr after head eversion (-4 AHE)], a stage prior to the onset of the ecdysone-triggered expression of target genes reaper and hid.
Figure 1.
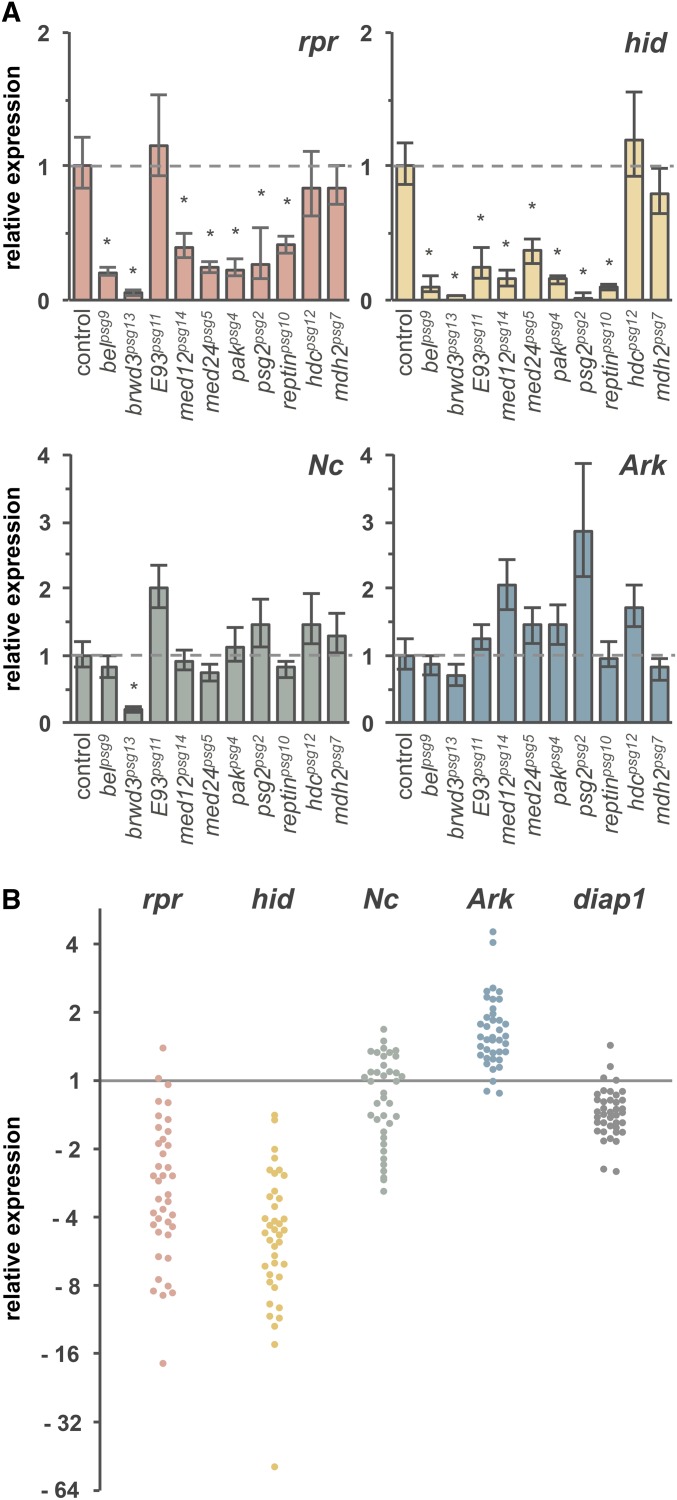
The ecdysone-induced expression of death activators is selectively disrupted in most PSG mutant salivary glands. (A) Eight of the 10 PSG mutations disrupt the ecdysone-induced expression of reaper (rpr) and/or hid in salivary glands dissected at 1.5 hr after head eversion (AHE). On the other hand, the ecdysone-induced expression of Ark and Nc is largely unaffected, indicating that the defects in hormone-induced transcription are target gene-specific. The y-axis plots relative expression of target genes measured by qPCR and compared to control. The x-axis shows control and mutant salivary glands. Each bar represents three independent biological samples normalized to rp49; asterisks indicate a significant reduction in expression compared to that in control glands (P-values <0.05). The allelic combinations used are shown in Table 1. (B) Molecular survey of 39 different PSG mutant salivary glands indicates that most PSG mutations disrupt initiation of the ecdysone-triggered death response. Expression of reaper, hid, Nc, Ark, and diap1 was measured in salivary glands dissected at +15 APF (equivalent to ∼ +3 AHE). The majority of PSG mutant glands display selective defects in the ecdysone-induced expression of rpr and hid, while Nc, Ark, and diap1 are expressed at near normal levels. Each dot represents a single sample of 30 salivary glands dissected from each PSG mutant analyzed. All samples were analyzed simultaneously and compared to three independent control samples dissected at +1.5 AHE (gray line at “1”). The y-axis shows relative expression (plotted on a log2 scale) normalized to both rpl18a and ubcd6.
Figure 6.
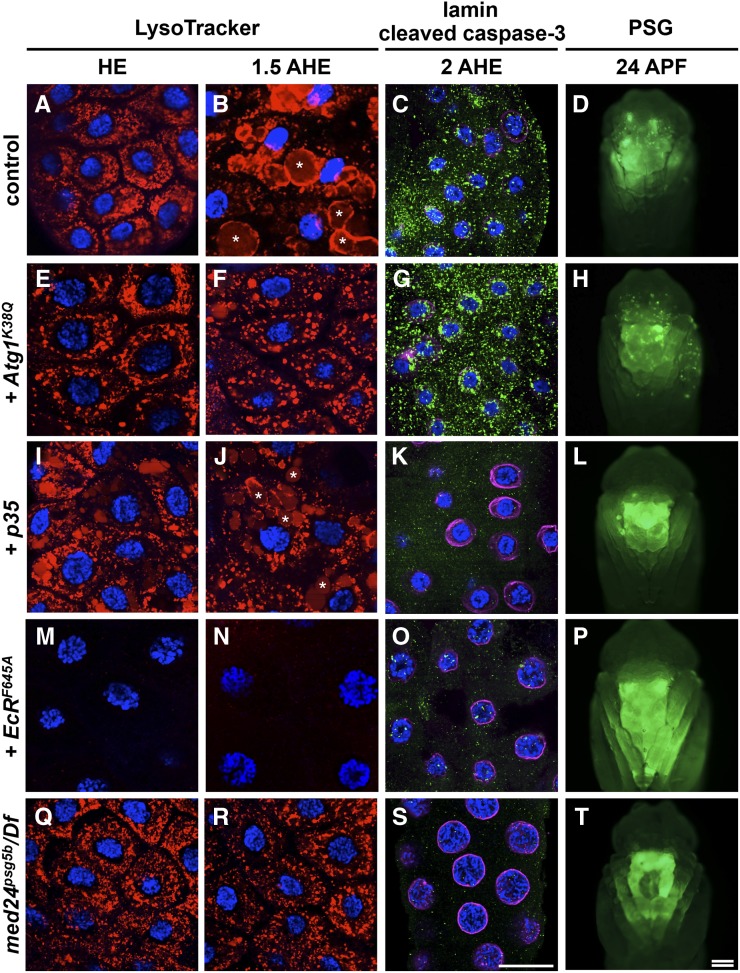
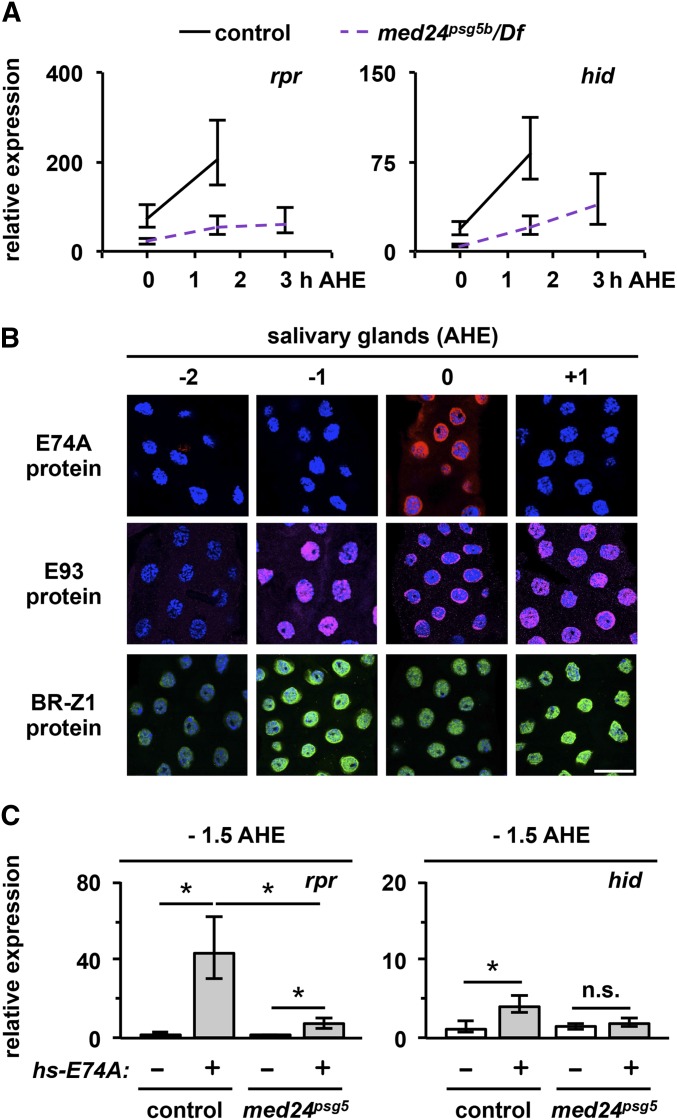
med24 is required for the ecdysone-triggered death response in larval salivary glands. (A–D) The death response in control salivary glands activates autophagy and caspases, resulting in tissue histolysis. LysoTracker staining (in red), used as a surrogate marker for autophagy, shows formation of large acidic structures only after the death response has been triggered (examples marked by white asterisks in B). In parallel, the death response triggers caspase activation (C), shown by staining with antibodies against cleaved caspase-3 (in green) and loss of staining of a target of caspases, nuclear lamin (in magenta). (D) The resulting tissue destruction is assayed by loss of the salivary gland-specific GFP expression (Sgs3-GAL4, UAS-GFP) at 24 hr after puparium formation (APF). (E–H) Salivary gland-specific expression of an inhibitor of autophagy, Atg1K38Q (UAS-Atg1K38Q/+; Sgs3-GAL4/+), blocks formation of the large LysoTracker-positive structures (E and F), but does not block caspase activation (G). Blocking only autophagy does not block tissue breakdown as shown by the absence of an intact PSG phenotype (H). (I–L) Conversely, expression of the caspase inhibitor p35 (UAS-p35/+; Sgs3-GAL4/+) does not block formation of large LysoTracker-positive structures (examples marked by white asterisks in J), but blocks activation of caspases (K). Blocking only caspase activation does not block tissue breakdown (L). (M–P) In contrast, blocking ecdysone signaling by expressing a dominant negative ecdysone receptor (UAS-EcRF645A/+; Sgs3-GAL4/+) blocks formation of any LysoTracker-positive structures (M and N) and blocks activation of caspases (O), resulting in a persistent salivary gland phenotype (P). (Q–T) In med24psg5 mutant glands, LysoTracker staining is similar to that in Atg1K38Q-expressing glands (Q and R); moreover, markers of caspase activity are similar to those of p35-expressing glands (S), suggesting that activation of autophagy and caspases does not occur. med24psg5 mutant animals have a PSG phenotype (T). Double bar in T, 200 μm for D, H, L, P, and T; bar in S, 50 μm for A–C, E–G, I–K, M–O, and Q–S. DAPI-costained nuclei are shown in blue. APF, hours after puparium formation; HE, head eversion; AHE, hours after head eversion; PSG, persistent salivary gland.
Immunofluorescence
Salivary glands were dissected from control and mutant animals staged relative to head eversion and stained with antibodies as previously described (Ihry et al. 2012). For cleaved caspase-3 staining, salivary glands were permeabilized by heptane treatment prior to fixation. The following primary antibodies were used: 1:50 mouse α-BR-C Z1 (Developmental Studies Hybridoma Bank), 1:50 mouse α-lamin (Developmental Studies Hybridoma Bank), 1:200 rabbit α-cleaved caspase-3 (Cell Signaling), 1:300 mouse α-E74A primary antibodies (a generous gift from C. Thummel), and 1:300 mouse α-E93 (a generous gift from I. Duncan; Washington University, Saint Louis MO). The following secondary antibodies were used at 1:200: Cy3 α-mouse and α-rabbit (Jackson Immuno-Research Laboratories) and AlexaFluor 488 and 633 α-mouse (Invitrogen). Stained salivary glands were mounted with Vectashield (Vector Laboratories, Burlingame, CA) and imaged on an Olympus FluoView FV1000 confocal microscope.
LysoTracker staining
Salivary glands dissected in PBS, from staged animals, were transferred to a 1.5-ml glass well plate containing a solution of LysoTracker Red at 50 nM and DAPI diluted 1:1000 (Invitrogen). Samples were covered and a rotator was used to agitate samples for 10 min at room temperature. Salivary glands were washed one time in PBS and mounted on glass slides in PBS to be imaged immediately by confocal microscopy.
Results
Mapping mutations with a PSG phenotype
As a first step toward functional characterization of the defects in mutant salivary glands, we mapped a subset of PSG complementation groups to their affected genes. We used recombination analysis with pairs of dominant markers and complementation tests with deficiencies to map 28 of the 37 PSG loci to discrete chromosomal regions (Sapiro et al. 2013). These 28 mutations were then tested for complementation with all publicly available lethal mutations within these mapped regions. The intent was to use an unbiased approach to rapidly identify a subset of PSG loci for further analysis. The PSG mutations identified in this manner map to the following 10 genes: belle (bel), brwd3/ram, E93 (Eip93F), headcase (hdc), mdh2, med12, med24, pak, psg2, and reptin (rept) (Table 1). Of these, the mapping of PSG mutations in belle (Ihry et al. 2012) and in med24, mdh2, and psg2 (Wang et al. 2008) have been reported. The remaining 19 PSG loci complemented all publicly available lethal mutations in their respective regions and were not pursued further.
All PSG mutations are lethal, with most animals arresting later during metamorphosis as pupae or pharate adults (Figure S1). Sanger sequencing shows that most of the newly mapped PSG alleles carry nonsense mutations that result in truncation of a significant portion of the encoded proteins (Table 1). Consistently, most mutations have similar lethal phases in homozygous and hemizygous (over a deficiency) combinations, suggesting that these mutations likely represent strong hypomorphic or loss-of-function alleles of the affected genes. On the other hand, hemizygous combinations of med12psg14 die as prepupae while those of brwd3psg13 and reptinpsg10 die earlier in development, suggesting that these PSG mutations are likely hypomorphic alleles. Our data also suggest that PSG mutations may represent unique alleles of their respective genes. For example, E93psg11 complements the E931 allele but fails to complement the E93vno allele, even though both lethal mutations map to the same gene (Mou et al. 2012). A similar allele-specific phenotype was recently described for belpsg9, which disrupts a subset of molecular functions regulated by belle (Ihry et al. 2012).
The PSG genes identified to date from this screen represent a functionally diverse group of proteins. belle is a DEAD box RNA-helicase required for translation of the ecdysone-induced transcription factor E74A (Ihry et al. 2012); in turn, E74A is required for the ecdysone-triggered destruction of salivary glands (Jiang et al. 2000; Wang et al. 2008). brwd3/ram contains a bromodomain and several WD40 repeats and likely regulates chromatin function (D’Costa et al. 2006). E93 encodes an ecdysone-induced transcription factor required for the destruction of salivary glands (Lee et al. 2000). hdc encodes an evolutionarily conserved protein containing domains with currently unknown molecular functions (Weaver and White 1995; Chien et al. 2006). mdh2 is a nuclear-encoded mitochondrial malate dehydrogenase (Wang et al. 2008, 2010). med12 and med24 are two subunits of the multiprotein mediator complex required for activator-dependent transcription (Malik and Roeder 2010). pak encodes a member of the highly conserved family of p21-activated kinases (Molli et al. 2009). psg2 encodes a protein product without defined molecular functions (Wang et al. 2008). And finally, reptin is an AAA+ family member with biochemical roles in both transcription and chromatin remodeling (Jha and Dutta 2009). Importantly, two of the mapped PSG genes, E93 and belle, encode regulators of ecdysone signaling required for the death response in salivary glands (Lee et al. 2000; Ihry et al. 2012). The roles of the remaining PSG genes in ecdysone signaling, if any, have not been previously characterized.
PSG mutations primarily disrupt the ability to initiate, rather than execute, the ecdysone-triggered death response
To begin to understand how PSG genes disrupt the death response in salivary glands, we measured the expression of ecdysone-induced target genes that initiate tissue destruction. The destruction of salivary glands requires activation of both caspases and autophagy (Berry and Baehrecke 2007). Although expression of several autophagy genes increases during the death response (Gorski et al. 2003; Lee et al. 2003), their role in activation of autophagic death and their relationship to ecdysone signaling have yet to be determined. In contrast, ecdysone activates caspases in salivary glands through transcriptional control of reaper and hid (Jiang et al. 1997, 2000). Because expression of reaper and hid is sufficient to initiate caspase activation (Grether et al. 1995; White et al. 1996), their induction defines a molecular readout, a point of no return, for the ecdysone-triggered death response. Failure to induce these genes, as occurs in E74A, br-Z1, or E93 mutant salivary glands (Jiang et al. 2000; Lee et al. 2002b), indicates defects upstream of the death response, likely within the ecdysone signaling cascade. Conversely, normal induction of reaper and hid may indicate defects downstream of the death response, possibly within the machinery that executes tissue destruction (e.g., caspases and/or autophagy). To measure whether reaper and hid were induced in response to ecdysone, we dissected salivary glands from control and mutant animals at +1.5 AHE, extracted total mRNA, and measured target gene expression using qPCR. This analysis was done at +1.5 AHE because reaper and hid are maximally induced in control glands at this stage. To our surprise, 8 of the 10 PSG mutations disrupt the ecdysone-dependent expression of these death activators (Figure 1A). Of these, only E93psg11 differentially affects death activator expression, disrupting hid but not reaper expression (the other 7 mutations disrupt expression of both reaper and hid; Figure 1A). The E93psg11 allele reveals a functional complexity within the E93 locus since the E931 allele disrupts expression of both reaper and hid (Lee et al. 2000, 2002b); this also suggests that induction of reaper and hid are genetically separable. Mutations in med24 have been previously reported to not affect reaper or hid expression (Wang et al. 2010), but these results were based on Northern blot analysis, which provides more qualitative than quantitative expression data. Importantly, despite the block in the ecdysone-triggered expression of reaper and hid, expression of other genes induced by the same pulse of ecdysone was not affected in these mutant glands. Ecdysone-induced expression of the caspase adaptor protein Ark (Lee et al. 2000; Ihry et al. 2012) is not disrupted in any mutant glands, while the ecdysone-induced expression of the initiator caspase Nc (Dorstyn et al. 1999; Lee et al. 2000) is disrupted only in brwd3psg13 mutant glands (Figure 1A). Although these results do not represent a comprehensive list of ecdysone-dependent targets in salivary glands, they do demonstrate that PSG mutations do not disable ecdysone signaling. Instead, most PSG mutations disrupt the expression of a subset of ecdysone-induced target genes, target genes like reaper and hid, which then activate pathways that execute tissue destruction.
To further demonstrate that defects upstream of the death response occur in most PSG mutations, we measured expression of death activators in salivary glands dissected from 39 different PSG mutant animals. These mutations represent the top 30 complementation groups identified in the original mutagenesis screen. For practical reasons, this molecular survey was performed using one biological replicate of 30 salivary glands dissected from each of the mutant animals. These results show that the defects described above are representative of the entire collection of PSG mutations. Expression of reaper and hid, but not Nc and Ark, was strongly disrupted in most PSG mutant glands (Figure 1B). In addition, the expression of the critical antiapoptotic gene, diap1, does not vary significantly among mutant glands, suggesting that the failure to initiate the death response is not caused by increased expression of apoptotic inhibitors. Despite the shortcomings of using one biological replicate, this molecular survey demonstrates that PSG mutations fall into two broad groups: those that disrupt the ability to initiate the death response (failing to induce reaper and hid) and those that disrupt the ability to execute the death response (inducing normal levels of reaper and hid). Most PSG mutations fall into the first group, those that disrupt the ability to initiate the death response.
PSG mutations disrupt a small subset of ecdysone-triggered responses during metamorphosis
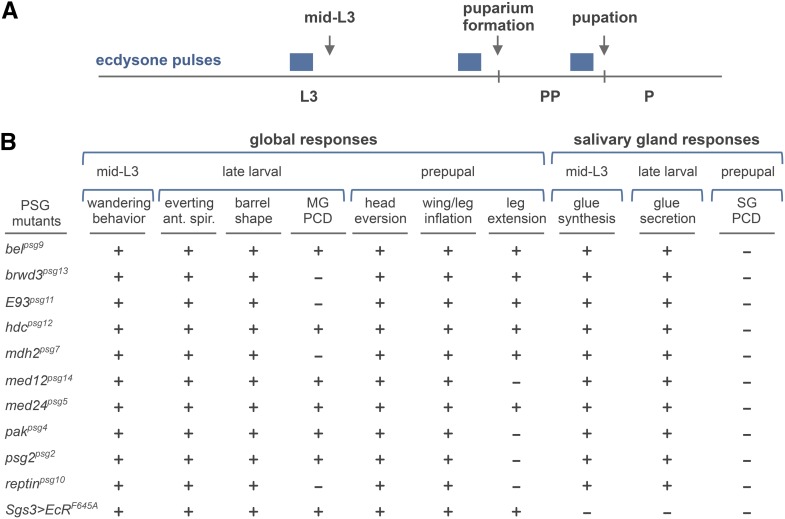
Given that most PSG mutations appear to disrupt ecdysone signaling during the death response in salivary glands, we examined whether these mutations were also required for other ecdysone-triggered responses at the onset of metamorphosis (Figure 2A). The mid-L3 pulse of ecdysone initiates larval wandering behavior (Riddiford 1993). About 24 hr later, the late larval pulse of ecdysone triggers eversion of the anterior spiracles and contraction of the larval body into the barrel shape of the future pupa (Ashburner 1989). This late larval pulse of ecdysone also initiates the destruction of larval midguts (Jiang et al. 1997; Lee et al. 2002a). Approximately 12 hr later, the prepupal pulse of ecdysone triggers eversion of the head capsule, as well as inflation of the wing and leg imaginal discs (Fristrom and Fristrom 1993). Subsequent extension of pupal legs is also thought to be dependent on ecdysone signaling (D’Avino and Thummel 1998). To our surprise, most of these stage-specific responses to ecdysone occur normally in PSG mutant animals (Figure 2B, Figure S2A). In fact, of all the ecdysone-triggered responses examined, PSG mutations disrupted, in addition to the destruction of larval salivary glands, only the destruction of larval midguts and the extension of leg imaginal discs. Importantly, however, the disruption of these two additional tissue-specific responses does not depend on the same set of PSG mutations, nor are they regulated by the same pulse of ecdysone, suggesting that each PSG gene regulates a small, but distinct, subset of ecdysone-triggered responses during metamorphosis.
Figure 2.
PSG genes are required for a subset of stage- and tissue-specific ecdysone-induced responses during the onset of metamorphosis. (A) Schematic showing the three systemic pulses of ecdysone (blue boxes) at the onset of metamorphosis. Arrows mark the major developmental transitions triggered by each of these pulses of ecdysone: the mid-third instar transition (mid-L3), puparium formation, and pupation. L3, third instar larvae; PP, prepupae; P, pupae. (B) Global and salivary gland-specific responses to each of the three pulses of ecdysone (referred to as the mid-L3, late larval, and prepupal pulses) were scored in PSG mutant animals. The “+” and “−” indicate whether or not appropriate responses were observed. Ant. spir, anterior spiracles; MG PCD, midgut programmed cell death; SG PCD, salivary gland programmed cell death. Genotype of Sgs3 > EcRF645A is UAS-EcRF645A/+; Sgs3-GAL4/+. Allelic combinations used are shown in Table 1.
The only apparent phenotype in common for all PSG mutations is the failure to destroy larval salivary glands (Figure 2B, Figure S2B). This tissue specificity may indicate a role for PSG genes during the development of salivary glands. To address this possibility, we examined salivary gland-specific responses to the same three pulses of ecdysone described above (Figure 2A). These pulses of ecdysone initiate distinct responses in developing salivary glands. The mid-L3 pulse of ecdysone initiates synthesis of glue proteins (Beckendorf and Kafatos 1976; Korge 1977); the late larval pulse initiates secretion of synthesized glue proteins (Biyasheva et al. 2001), allowing newly-formed pupae to stick to solid surfaces; and, finally, the prepupal pulse of ecdysone triggers the death response (Jiang et al. 1997). Synthesis of glue proteins was examined using qPCR to measure ecdysone-dependent induction of the glue gene Sgs3 (Figure 2B, Figure S3) and secretion of glue proteins was examined by the ability of pupae to firmly adhere to the sides of vials (Figure 2B). As expected, all of these responses to ecdysone were blocked by salivary gland-specific expression of a dominant negative version of the ecdysone receptor (EcRF645A; Figure 2B, Figure S3). PSG mutant animals, however, disrupted only the death response, not the responses to the prior two pulses of ecdysone (Figure 2B, Figure S3), suggesting that PSG mutations do not disrupt the overall development of salivary glands. Taken together, the PSG mutations encode genes that regulate a highly-selective subset of stage- and tissue-specific biological responses initiated by systemic pulses of ecdysone.
Most PSG genes are induced in a stage- and tissue-specific manner during the ecdysone-triggered death response
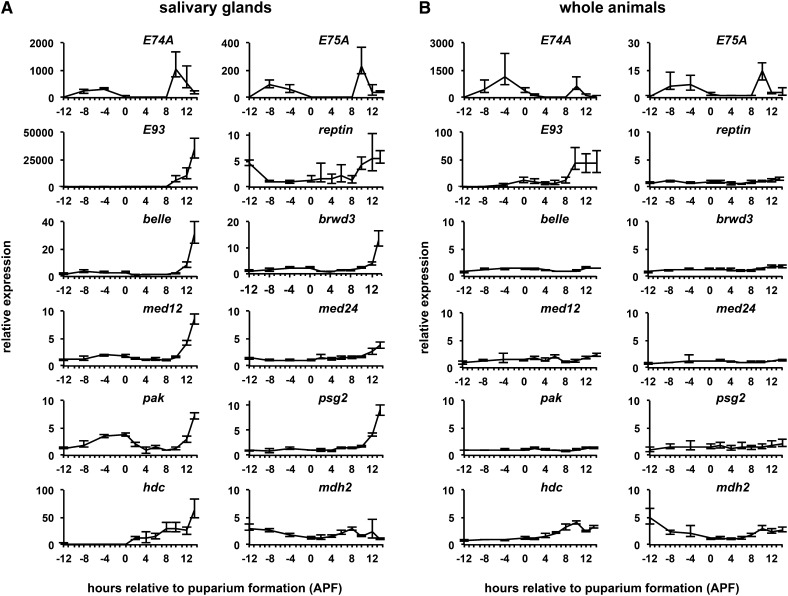
To understand how PSG mutations regulate the death response in such a specific manner, we examined when and where these PSG genes were expressed. Total mRNA was extracted from either dissected salivary glands or whole animals at various developmental stages from early wandering third instar larvae until after the death response (−12 APF through +13.5 APF). The two systemic pulses of ecdysone that occur during this period, the late larval pulse at −4 APF and the prepupal pulse at +10 APF, are highlighted by the induction of the ecdysone primary response genes E74A and E75A (Figure 3A). Consistent with published reports (Baehrecke and Thummel 1995), E93 showed a strong stage-specific induction in response to the prepupal pulse of ecdysone in salivary glands (Figure 3A). Strikingly, most PSG genes also showed a stage-specific expression profile in salivary glands similar to that of E93. belle, brwd3, med12, pak, psg2, and, to a lesser but statistically significant extent, med24 and rept, show increased expression during the death response (Figure 3A). Moreover, these stage-specific expression profiles were also tissue-specific: whole animal expression profiles of these genes, other than that of E93, are relatively flat throughout the stages examined (Figure 3B). In contrast, the two remaining PSG genes, hdc and mdh2, have different expression profiles (Figure 3). hdc shows a gradual increase in expression after puparium formation in glands and in whole animals, with a spike in expression immediately after head eversion in salivary glands. mdh2 remains relatively flat throughout this period in both expression profiles. Thus, most PSG genes are induced in a stage- and tissue-specific manner during the ecdysone-triggered death response in salivary glands.
Figure 3.
Most PSG genes show stage- and tissue-specific expression profiles. (A and B) Developmental expression profiles of target genes measured by qPCR in dissected salivary glands (A) or in whole animals (B). Previously characterized ecdysone target genes E74A and E75A show strong induction in response to the late larval (at −4 APF) and prepupal (at +10 APF) pulses of ecdysone, while E93 responds only to the prepupal pulse. Most PSG genes, however, show strong induction after the prepupal pulse of ecdysone in salivary glands yet remain relatively flat in whole animals. The y-axis plots relative expression measured by qPCR for each target gene compared to the stage with the lowest expression. The x-axis plots developmental stage relative to puparium formation in control salivary glands and whole animals. Each time point represents three independent biological samples normalized to rp49.
PSG genes brwd3, med12, med24, pak, and psg2 encode novel ecdysone secondary response genes
The expression profiles of most PSG genes suggest that they may be induced by the prepupal pulse of ecdysone. In fact, one of the best-characterized stage-specific targets of ecdysone is E93 (Baehrecke and Thummel 1995; Lee et al. 2000). E93 is an ecdysone primary response gene, directly induced by the prepupal pulse of ecdysone, and thought to provide specificity to ecdysone responses (Lee et al. 2000; Mou et al. 2012). To determine whether the remaining PSG genes were also targets of ecdysone, we first tested whether ecdysone was required for expression of these genes. Indeed, blocking ecdysone signaling immediately prior to the prepupal pulse of ecdysone, with EcRF645A, disrupts the salivary gland-specific induction of all PSG genes except reptin (Figure 4A). As expected, expression of mdh2, which is not upregulated during this stage, is also unaffected by EcRF645A (Figure 4A). We then tested whether these newly-identified genes were primary or secondary response genes within the ecdysone transcriptional hierarchy. To do this, we measured expression of the ecdysone-dependent PSG genes in salivary glands dissected at +1.5 AHE from animals carrying null mutations in the ecdysone primary response genes E74A (E74Aneo24), E93 (E931), and br-Z1 (brrbp5). Ecdysone primary response genes are directly induced by ecdysone and their induction does not depend on activity of other primary response genes. In contrast, expression of the newly-identified ecdysone-dependent genes is regulated by primary response genes. Six of the PSG genes (belle, brwd3, med12, med24, pak, and psg2) require all three ecdysone primary response genes for their expression during the death response (Figure 4B). Together with expression profiles that coincide with ecdysone pulses (Figure 3A), these six genes fit the classical definition of ecdysone secondary response genes (Ashburner 1974). On the other hand, expression of hdc, although dependent on a subset of primary response genes (Figure 4B), does not display a classical ecdysone-dependent expression profile (Figure 3A).
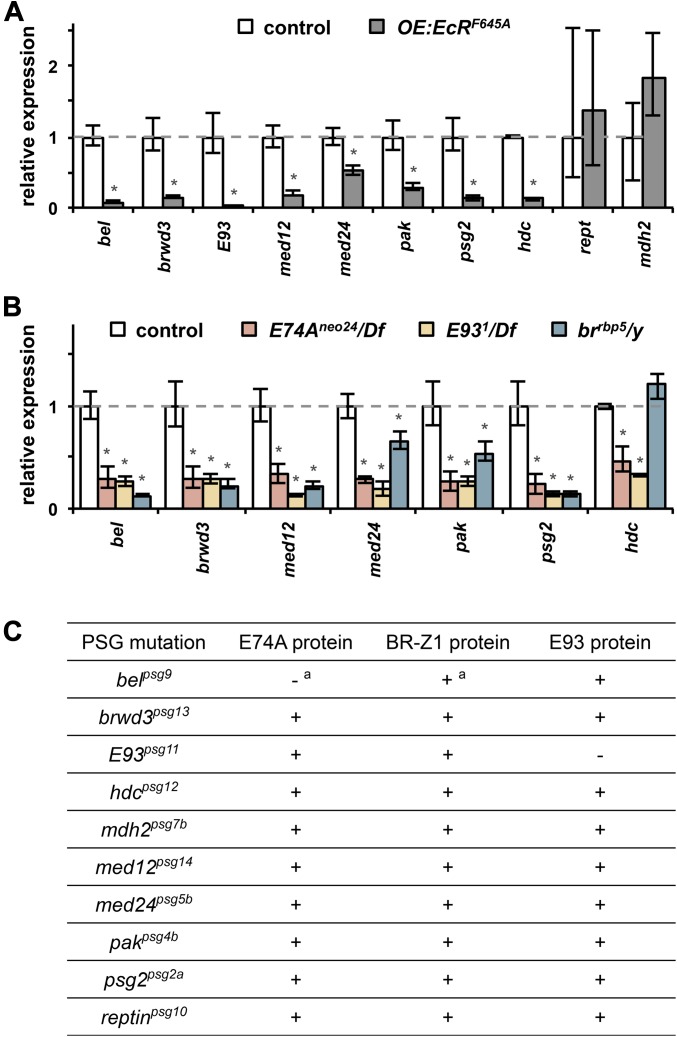
Figure 4.
Many PSG genes encode components of the ecdysone-triggered transcriptional hierarchy. (A) Expression of a dominant negative version of the ecdysone receptor (EcRF645A) prior to the prepupal pulse of ecdysone blocks expression of most PSG genes in salivary glands, showing that the stage-specific induction in salivary glands is hormone-dependent. Control salivary glands (white bars) were dissected 1.5 hr after head eversion (AHE). OE:EcRF645A (UAS-GAL4/UAS-EcRF645A; hs-GAL4/+) glands (gray bars) were dissected at +1.5 AHE, after prepupae were heat-shocked 4 hr before head eversion (−4 AHE). The x-axis shows all 10 PSG genes and the y-axis plots relative expression compared to control and normalized to rp49. (B) The ecdysone primary response genes E74A, E93, and br-Z1 are required for the ecdysone-dependent induction of PSG genes in salivary glands. Shown is qPCR analysis of PSG gene expression in salivary glands dissected from control and mutant animals at +1.5 AHE. The x-axis shows all ecdysone-dependent PSG genes (except E93, which is a known ecdysone primary response gene). The colors of the bars indicate the genotype of the dissected salivary glands at +1.5 AHE: control (white), E74Aneo24/Df (pink), E931/Df (yellow), or brrbp5/y (blue). The y-axis plots relative expression of target genes compared to control and normalized to rp49. All qPCR data represent results from three independent biological samples with asterisks indicating significant change in expression compared to that in control glands (P-values <0.05). (C) Translation of ecdysone primary response proteins E74A, E93, and BR-Z1 in PSG mutant salivary glands. All mutant glands show robust expression of the three primary response genes (marked by “+”), except for absence of E74A protein in belpsg9 and E93 protein in E93psg11. Salivary glands were dissected at head eversion and analyzed by immunofluorescence with antibodies directed to E74A, E93, and BR-Z1 proteins. a, previously reported in Ihry et al. (2012).
We have recently shown that the PSG gene belle is critical for translation of E74A mRNA (Ihry et al. 2012). Here we show that induction of belle during the death response depends on the primary response gene it regulates, demonstrating cross-talk between layers of the ecdysone-triggered transcriptional hierarchy, whereby an ecdysone secondary response gene is required for translational control of an ecdysone primary response gene. Although the functional significance of this cross-talk has yet to be determined, it raises the possibility that other PSG genes may also disrupt translation of ecdysone primary response genes. To examine this possibility, we stained salivary glands, dissected at head eversion, with antibodies directed to E74A, E93, and BR-Z1 proteins. These three proteins are the only ecdysone primary response genes known to be required for the death response in salivary glands (Jiang et al. 2000; Lee et al. 2002b). As expected, E74A protein was absent in belpsg9 mutant glands, and E93 protein was absent in E93psg11 mutant glands (Figure 4C). However, all the remaining combinations were positive (Figure 4C), demonstrating that PSG genes, with the exception of belle and E93, do not disrupt translation of ecdysone primary response genes. Taken together, these results indicate that half of the PSG genes, brwd3, med12, med24, pak, and psg2, encode bona fide secondary response genes within the ecdysone transcriptional hierarchy.
The ecdysone secondary response gene med24 is required to amplify target gene expression during the death response
Our results indicate that all of the newly-identified ecdysone secondary response genes are required for the ecdysone-triggered death response, disrupting the ecdysone-triggered expression of reaper and hid. However, the role of these secondary response genes during ecdysone signaling remains unclear. To address this problem, we focused on one of the ecdysone secondary response genes, med24. Hemizygous med24psg5 mutant animals have one of the most highly penetrant PSG phenotypes (Figure S2B), making it an ideal candidate for further analysis. We first examined the developmental expression profiles of reaper and hid in control and mutant salivary glands. In control glands, induction of reaper and hid starts at head eversion in response to the prepupal pulse of ecdysone and continues to increase thereafter (Figure 5A). In med24psg5 glands, induction of reaper also starts at head eversion but at three times lower levels: 23-fold induction in med24psg5 compared to 71-fold induction in control glands (Figure 5A). Although the expression of reaper increases thereafter in mutant glands, it remains significantly lower than in control glands. The expression of hid shows a similarly submaximal expression profile in mutant glands (Figure 5A). In contrast to these results, expression of EcRF645A results in a complete disruption of reaper and hid expression (Ihry et al. 2012), suggesting that within the ecdysone transcriptional hierarchy med24psg5 regulates the amplitude, not the onset, of the ecdysone-induced transcriptional response. This defect is consistent with the observed submaximal expression of reaper and hid in all PSG mutations that disrupt the death response (Figure 1A) and reflects the biological significance of inducing sufficiently high levels of these death activators (Yin and Thummel 2004).
Figure 5.
The PSG gene med24 amplifies hormone-dependent expression of effector genes reaper and hid. (A) Developmental expression profiles of rpr or hid in control (black solid line) and med24 mutant (purple dashed line) salivary glands. rpr and hid are induced, but at submaximal levels in mutant glands. The y-axis plots relative expression of reaper or hid as measured by qPCR. The expression ratios were calculated relative to those in −4 AHE control salivary glands, a stage prior to the prepupal pulse of ecdysone and the onset of reaper and hid expression. The x-axis plots developmental stage relative to hours after head eversion (AHE). (B) E74A protein expression coincides with the onset of rpr and hid transcription at head eversion, while E93 and Z1 proteins are robustly expressed earlier. Salivary glands were dissected and analyzed by immunofluorescence for expression of E74A (red), E93 (magenta), and BR-Z1 (green) proteins. Nuclei were costained with DAPI (blue). Bar, 50 μm. (C) Precocious expression of E74A protein is sufficient to induce expression of reaper and hid. Although ectopic E74A can induce reaper in med24 mutant glands, this induction is significantly lower than the response observed in control glands. Ectopic E74A cannot induce hid in med24 mutant glands. Control and mutant animals carrying one copy of the hs-E74A transgene (gray bars) were heat-shocked for 30 min at −4 AHE and salivary glands dissected 2 hr later (at −1.5 AHE). The same heat-shock treatment was given to control and mutant glands without the hs-E74A transgene (white bars). The x-axis shows the genotypes and the presence (+) or absence (−) of the hs-E74A transgene. The y-axis plots relative expression of reaper or hid as measured by qPCR. The expression ratios were calculated relative to those in −4 AHE control salivary glands. All qPCR data represent three independent salivary gland samples normalized to rp49; asterisks indicate significant differences in expression (P-values <0.05).
Defects in amplitude of transcriptional responses reflect a slower rate of transcriptional initiation; however, defects in the rate of transcription are often difficult to distinguish from defects in the timing of transcription. This is important for hormonal signaling cascades because, for example, a relatively minor delay in the timing of the hormone pulse may result in significantly lower levels of accumulated transcripts. To separate defects in amplitude and timing, we first identified a critical regulator of the timing of the death response downstream of ecdysone. Ecdysone primary response genes E74A, E93, and br-Z1 are coordinately transcribed in response to the prepupal pulse of ecdysone (Thummel et al. 1990; Karim and Thummel 1992; Baehrecke and Thummel 1995). Their protein products, however, are translated at different times. Using antibodies directed to the E74A, E93, and BR-Z1 proteins, we show that translation of E74A protein coincides with head eversion, while translation of E93 and BR-Z1 proteins occurs at least 1 hr earlier (Figure 5B). This result raises the possibility that translation of E74A protein may determine the timing of the death response. Accordingly, precocious expression of E74A protein from a heterologous promoter (Ihry et al. 2012), ∼1.5 hr before head eversion (−1.5 AHE), is sufficient to trigger an ∼44-fold induction of reaper expression (Figure 5C). On the other hand, the same treatment in med24psg5 mutant glands triggered an ∼14-fold induction of reaper (Figure 5C), three times lower than the induction observed in control glands. Precocious E74A protein had a smaller, but statistically significant, effect on expression of hid in control salivary glands (Figure 5C). This E74A-dependent response of hid is absent in med24psg5 mutant glands (Figure 5C). Taken together, these results suggest that med24, and most likely the other ecdysone secondary genes, regulate the maximal induction of reaper and hid, consistent with a role in amplification during hormone-dependent transcription.
med24 is required to initiate tissue destruction in salivary glands
To demonstrate that the defects in amplification of the death response result in the subsequent failure to execute tissue destruction, we stained med24psg5 mutant glands for markers of caspase activation and autophagy. Phenotypic analysis of autophagy is difficult and labor-intensive, often requiring electron microscopy to evaluate the number and size of vesicles during autophagosome formation (Klionsky et al. 2012). We used LysoTracker as a potential surrogate for autophagy since this vital dye stains all acidic structures within cells. At head eversion, salivary gland cells are filled with small LysoTracker-positive acidic structures (Figure 6A). About 1.5 hr after head eversion (+1.5 AHE), after the death response has initiated, LysoTracker stains much larger structures (note white asterisks in Figure 6B). Expression of the inhibitor of autophagy Atg1K38Q, however, blocks formation of these large acidic structures (Figure 6, E and F). In contrast, expression of the apoptotic inhibitor p35 does not disrupt formation of large LysoTracker-positive structures (Figure 6, I and J). Conversely, expression of p35, but not Atg1K38Q, disrupts the activation of caspases during the death response in salivary glands. This is shown by staining with antibodies that detect activity of the initiator caspase Nc (Fan and Bergmann 2009) and nuclear lamin, a direct target of caspases (Figure 6, C, G, and K).
Disrupting ecdysone signaling by expression of EcRF645A blocks activation of caspases and formation of any acidic structures (Figure 6, M–O), resulting in a strong PSG phenotype (Figure 6P). On the other hand, blocking autophagy or apoptosis alone does not result in a strong PSG phenotype (Figure 6, H and L). The persistent GFP fragments likely reflect slower tissue breakdown when either caspases or autophagy are blocked. Accordingly, the salivary glands in these animals are too fragile to dissect at +24 APF (Figure S4). In contrast, EcRF645A-expressing glands can be easily dissected at +24 APF (Figure S4), a faithful measure of the failure in tissue histolysis. Importantly, consistent with its role within ecdysone signaling, med24psg25 mutant salivary glands disrupt caspase activation and the formation of large LysoTracker-positive structures (Figure 6, Q–S), resulting in a strong PSG phenotype with glands that can be dissected at +24 APF (Figure 6T, Figure S4). med24 was previously reported not to affect autophagy (Wang et al. 2010); however, that conclusion was based solely on the presence of LysoTracker staining. Our analysis demonstrates that med24 mutant glands, like Atg1K38Q-expressing glands, block formation of the larger acidic structures (Figure 6, F and R), suggesting a possible influence of med24 on autophagy. Although the ecdysone-dependent target genes that initiate autophagy during the death response have yet to be identified, our results provide genetic evidence that the activation of autophagy and caspases is likely regulated by a common set of genes within the ecdysone transcriptional hierarchy.
Discussion
Systemically-released pulses of steroid hormones initiate a wide variety of biological responses during development and homeostasis. Our results demonstrate that components within the steroid-triggered transcriptional hierarchy itself play a critical role in refining these systemic signals into distinct local responses. These components include novel ecdysone secondary response genes; genes like brwd3, med12, med24, pak, and psg2 that are themselves induced by ecdysone in a stage- and tissue-specific manner. These ecdysone secondary response genes help amplify the hormonal cue, but do so in a tissue-, stage-, and target-specific manner. The result is a dramatic, hormone-dependent, transcriptional activation of genes that initiate the appropriate biological responses.
The experimental context for this study is the ecdysone-triggered destruction of salivary glands during metamorphosis. The original motivation in screening for mutations that disrupt this process was to identify novel regulators of cell death (Wang et al. 2008). However, about half of the pupal lethal mutations examined during the mutagenesis screen had a persistent salivary gland phenotype (Wang et al. 2008). To winnow this common phenotype, we selected only those mutations with “intact” salivary glands inside pupae with relatively normal morphology. By adding this criterion, we had hoped to select against mutations in global regulators of ecdysone signaling. In hindsight, however, it appears we have also selected against mutations in regulators of caspase activation or autophagy because individually blocking these processes results in incomplete or partial destruction of salivary glands (Leulier et al. 2006; Berry and Baehrecke 2007). In fact, an independent screen for suppressors of ectopic reaper in the eye [Su(GMR-rpr)] identified a gain-of-function mutation in diap1 as well as two loss-of-function mutations in the caspase Nc, all present among the original, but “non-PSG”, pupal lethal mutations (A. Bashirullah, unpublished data). Nevertheless, a number of mutations within our collection of PSG mutations, like those in hdc and mdh2, may provide insights into the mechanisms that execute tissue destruction. mdh2 is required to generate sufficient ATP, a key and rate-limiting ingredient during tissue destruction (Wang et al. 2010). The role of hdc during tissue destruction, however, remains to be determined. Importantly, our results demonstrate that the selection used to identify PSG phenotypes has primarily enriched for mutations that disrupt the initiation, rather than the execution, of the death response, highlighting the role of the ecdysone-triggered transcriptional hierarchy during this stage- and tissue-specific biological response.
Our data suggest that many, if not most, PSG mutations disrupting ecdysone signaling within salivary glands encode components of the ecdysone-triggered transcriptional hierarchy. All of these genes are induced in a stage- and tissue-specific manner by ecdysone and they are, in turn, required for induction of a subset of ecdysone-dependent target genes. These newly-identified components of the ecdysone transcriptional hierarchy greatly expand the number of genes known to provide specificity to ecdysone pulses during development. Strikingly, most of these components of the ecdysone signaling hierarchy encode Drosophila homologs of vertebrate nuclear receptor coregulators. Both Med12 and Med24 bind to and are required for function of a large number of nuclear receptors, including the thyroid receptor (Ito et al. 1999; Zhang and Fondell 1999; Fondell 2013), the androgen receptor (Wang et al. 2002), the estrogen receptor (Kang et al. 2002), and the vitamin D receptor (Ito et al. 1999; Zhang and Fondell 1999). One of the pak homologs in vertebrates, pak6, can directly regulate androgen and estrogen receptor function (Yang et al. 2001; Lee et al. 2002). In addition, the mammalian ortholog of E93 is the ligand-dependent corepressor LCoR that is required for transcriptional activation by the estrogen, androgen, and progesterone receptors (White et al. 2004; Palijan et al. 2009; Asim et al. 2011). Moreover, although nuclear receptor coactivator functions have not been reported for human brwd3, recent proteomic studies have reported interaction of the human BRWD3 with nuclear receptors (Miyamoto-Sato et al. 2010).
Our results highlight important parallels between the role of these newly-identified ecdysone response genes in insects and those of nuclear receptor coregulators in vertebrates. Both appear to be required for amplification, but not initiation, of hormone-dependent transcription (Mckenna and O’Malley 2002). Moreover, there is evidence that vertebrate coregulators may also be targets of the hormonal responses they amplify. For example, expression of a subset of androgen receptor coregulators can be upregulated by androgens in human cell lines (Urbanucci et al. 2008). Given that increased expression levels of nuclear receptor coactivators generate increased hormone-dependent transcription (Chen et al. 2000), the relationship between coactivator function and their increased expression is likely to confer a faster and stronger hormonal response. Moreover, in addition to this hierarchical relationship between hormones and their coregulators, our results also highlight a combinatorial relationship between hormones and their coregulators. Like vertebrate coregulators, ecdysone secondary response genes appear to play a critical role in providing specificity to systemic hormonal pulses, whereby each coactivator regulates a small, but largely nonoverlapping, subset of steroid-triggered biological responses. These results suggest that the same coregulators may be used in different combinations in different tissues to regulate distinct biological processes.
Improper function or expression of nuclear receptor coregulators has been implicated in a rapidly growing list of human diseases (Lonard and O’Malley 2012). However, the number of human nuclear receptor coregulators estimated by proteomics may be in the hundreds and even thousands (Malovannaya et al. 2011; O’Malley et al. 2012), making it difficult to identify those that play more important roles in development and disease. The degree of functional conservation in the regulation of nuclear receptor signaling described here suggests that forward genetic approaches in Drosophila may help identify novel and clinically relevant coregulators of steroid hormone signaling. Accordingly, the human homologs of five of the six components of ecdysone signaling we identified in this genetic screen have been directly linked to endocrine-based disorders like breast, prostate, and ovarian cancers (Kim et al. 2005; Siu et al. 2010; Asim et al. 2011; Barbieri et al. 2012; Hasegawa et al. 2012).
Supplementary Material
Acknowledgments
We thank the Bloomington Drosophila Stock Center, E. Baehrecke, K. Moses, and C. Thummel for stocks and Yunsik Kang and Nick Wleklinski for technical assistance. This work has been supported in part by a National Institute of General Medical Sciences/National Institutes of Health grant (R01 GM095944) (to A.B.).
Footnotes
Communicating editor: M. Wolfner
Literature Cited
- Andres A. J., Thummel C. S., 1994. Methods for quantitative analysis of transcription in larvae and prepupae. Methods Cell Biol. 44: 565–573. [DOI] [PubMed] [Google Scholar]
- Ashburner M., 1974. Sequential gene activation by ecdysone in polytene chromosomes of Drosophila melanogaster. II. The effects of inhibitors of protein synthesis. Dev. Biol. 39: 141–157. [DOI] [PubMed] [Google Scholar]
- Ashburner M., 1989. Drosophila: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Ashburner M., Chihara C., Meltzer P., Richards G., 1974. Temporal control of puffing activity in polytene chromosomes. Cold Spring Harb. Symp. Quant. Biol. 38: 655–662. [DOI] [PubMed] [Google Scholar]
- Asim M., Hafeez B. B., Siddiqui I. A., Gerlach C., Patz M., et al. , 2011. Ligand-dependent corepressor acts as a novel androgen receptor corepressor, inhibits prostate cancer growth, and is functionally inactivated by the Src protein kinase. J. Biol. Chem. 286: 37108–37117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baehrecke E. H., Thummel C. S., 1995. The Drosophila E93 gene from the 93F early puff displays stage- and tissue-specific regulation by 20-hydroxyecdysone. Dev. Biol. 171: 85–97. [DOI] [PubMed] [Google Scholar]
- Bainbridge S. P., Bownes M., 1981. Staging the metamorphosis of Drosophila melanogaster. J. Embryol. Exp. Morphol. 66: 57–80. [PubMed] [Google Scholar]
- Barbieri C. E., Baca S. C., Lawrence M. S., Demichelis F., Blattner M., et al. , 2012. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat. Genet. 44: 685–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckendorf S. K., Kafatos F. C., 1976. Differentiation in the salivary glands of Drosophila melanogaster: characterization of the glue proteins and their developmental appearance. Cell 9: 365–373. [DOI] [PubMed] [Google Scholar]
- Berry D. L., Baehrecke E. H., 2007. Growth arrest and autophagy are required for salivary gland cell degradation in Drosophila. Cell 131: 1137–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biyasheva A., Do T. V., Lu Y., Vaskova M., Andres A. J., 2001. Glue secretion in the Drosophila salivary gland: a model for steroid-regulated exocytosis. Dev. Biol. 231: 234–251. [DOI] [PubMed] [Google Scholar]
- Chen S., Sarlis N. J., Simons S. S., 2000. Evidence for a common step in three different processes for modulating the kinetic properties of glucocorticoid receptor-induced gene transcription. J. Biol. Chem. 275: 30106–30117. [DOI] [PubMed] [Google Scholar]
- Chien C.-C., Chang C.-C., Yang S.-H., Chen S.-H., Huang C.-J., 2006. A homologue of the Drosophila headcase protein is a novel tumor marker for early-stage colorectal cancer. Oncol. Rep. 15: 919–926. [PubMed] [Google Scholar]
- D’Avino P. P., Thummel C. S., 1998. Crooked legs encodes a family of zinc finger proteins required for leg morphogenesis and ecdysone-regulated gene expression during Drosophila metamorphosis. Development 125: 1733–1745. [DOI] [PubMed] [Google Scholar]
- D’Costa A., Reifegerste R., Sierra S., Moses K., 2006. The Drosophila ramshackle gene encodes a chromatin-associated protein required for cell morphology in the developing eye. Mech. Dev. 123: 591–604. [DOI] [PubMed] [Google Scholar]
- Dorstyn L., Colussi P. A., Quinn L. M., Richardson H., Kumar S., 1999. DRONC, an ecdysone-inducible Drosophila caspase. Proc. Natl. Acad. Sci. USA 96: 4307–4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y., Bergmann A., 2009. The cleaved-Caspase-3 antibody is a marker of Caspase-9-like DRONC activity in Drosophila. Cell Death Differ. 17: 534–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fondell J. D., 2013. The Mediator complex in thyroid hormone receptor action. Biochim. Biophys. Acta 1830: 3867–3875. [DOI] [PubMed] [Google Scholar]
- Fristrom D., Fristrom J. W., 1993. The metamorphic development of the adult epidermis, pp. 843–897 in The Development of Drosophila melanogaster, edited by Bate M., Martinez-Arias A. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Gorski S. M., Chittaranjan S., Pleasance E. D., Freeman J. D., Anderson C. L., et al. , 2003. A SAGE approach to discovery of genes involved in autophagic cell death. Curr. Biol. 13: 358–363. [DOI] [PubMed] [Google Scholar]
- Grether M. E., Abrams J. M., Agapite J., White K., Steller H., 1995. The head involution defective gene of Drosophila melanogaster functions in programmed cell death. Genes Dev. 9: 1694–1708. [DOI] [PubMed] [Google Scholar]
- Hasegawa N., Sumitomo A., Fujita A., Aritome N., Mizuta S., et al. , 2012. Mediator subunits MED1 and MED24 cooperatively contribute to pubertal mammary gland development and growth of breast carcinoma cells. Mol. Cell. Biol. 32: 1483–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihry R. J., Sapiro A. L., Bashirullah A., 2012. Translational control by the DEAD box RNA helicase belle regulates ecdysone-triggered transcriptional cascades. PLoS Genet. 8: e1003085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito M., Yuan C.-X., Malik S., Gu W., Fondell J. D., et al. , 1999. Identity between TRAP and SMCC complexes indicates novel pathways for the function of nuclear receptors and diverse mammalian activators. Mol. Cell 3: 361–370. [DOI] [PubMed] [Google Scholar]
- Jha S., Dutta A., 2009. RVB1/RVB2: running rings around molecular biology. Mol. Cell 34: 521–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C., Baehrecke E. H., Thummel C. S., 1997. Steroid regulated programmed cell death during Drosophila metamorphosis. Development 124: 4673–4683. [DOI] [PubMed] [Google Scholar]
- Jiang C., Lamblin A. F., Steller H., Thummel C. S., 2000. A steroid-triggered transcriptional hierarchy controls salivary gland cell death during Drosophila metamorphosis. Mol. Cell 5: 445–455. [DOI] [PubMed] [Google Scholar]
- Kang Y. K., Guermah M., Yuan C.-X., Roeder R. G., 2002. The TRAP/Mediator coactivator complex interacts directly with estrogen receptors alpha and beta through the TRAP220 subunit and directly enhances estrogen receptor function in vitro. Proc. Natl. Acad. Sci. USA 99: 2642–2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karim F. D., Thummel C. S., 1992. Temporal coordination of regulatory gene expression by the steroid hormone ecdysone. EMBO J. 11: 4083–4093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. H., Kim B., Cai L., Choi H. J., Ohgi K. A., et al. , 2005. Transcriptional regulation of a metastasis suppressor gene by Tip60 and beta-catenin complexes. Nature 434: 921–926. [DOI] [PubMed] [Google Scholar]
- Klionsky D. J., Abdalla F. C., Abeliovich H., Abraham R. T., Acevedo-Arozena A., et al. , 2012. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy 8: 445–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korge G., 1977. Larval saliva in Drosophila melanogaster: production, composition, and relationship to chromosome puffs. Dev. Biol. 58: 339–355. [DOI] [PubMed] [Google Scholar]
- Lee C. Y., Baehrecke E. H., 2001. Steroid regulation of autophagic programmed cell death during development. Development 128: 1443–1455. [DOI] [PubMed] [Google Scholar]
- Lee C. Y., Wendel D. P., Reid P., Lam G., Thummel C. S., et al. , 2000. E93 directs steroid-triggered programmed cell death in Drosophila. Mol. Cell 6: 433–443. [DOI] [PubMed] [Google Scholar]
- Lee C.-Y., Cooksey B. A. K., Baehrecke E. H., 2002a Steroid regulation of midgut cell death during Drosophila development. Dev. Biol. 250: 101–111. [DOI] [PubMed] [Google Scholar]
- Lee C.-Y., Simon C. R., Woodard C. T., Baehrecke E. H., 2002b Genetic mechanism for the stage- and tissue-specific regulation of steroid triggered programmed cell death in Drosophila. Dev. Biol. 252: 138–148. [DOI] [PubMed] [Google Scholar]
- Lee C.-Y., Clough E. A., Yellon P., Teslovich T. M., Stephan D. A., et al. , 2003. Genome-wide analyses of steroid- and radiation-triggered programmed cell death in Drosophila. Curr. Biol. 13: 350–357. [DOI] [PubMed] [Google Scholar]
- Lee S. R., Ramos S. M., Ko A., Masiello D., Swanson K. D., et al. , 2002. AR and ER interaction with a p21-activated kinase (PAK6). Mol. Endocrinol. 16: 85–99. [DOI] [PubMed] [Google Scholar]
- Leulier F., Ribeiro P. S., Palmer E., Tenev T., Takahashi K., et al. , 2006. Systematic in vivo RNAi analysis of putative components of the Drosophila cell death machinery. Cell Death Differ. 13: 1663–1674. [DOI] [PubMed] [Google Scholar]
- Lonard D. M., O’Malley B. W., 2007. Nuclear receptor coregulators: judges, juries, and executioners of cellular regulation. Mol. Cell 27: 691–700. [DOI] [PubMed] [Google Scholar]
- Lonard D. M., O’Malley B. W., 2012. Nuclear receptor coregulators: modulators of pathology and therapeutic targets. Nat. Rev. Endocrinol. 8: 598–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik S., Roeder R. G., 2010. The metazoan Mediator co-activator complex as an integrative hub for transcriptional regulation. Nat. Rev. Genet. 11: 761–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malovannaya A., Lanz R. B., Jung S. Y., Bulynko Y., Le N. T., et al. , 2011. Analysis of the human endogenous coregulator complexome. Cell 145: 787–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mckenna N. J., O’Malley B. W., 2002. Combinatorial control of gene expression by nuclear receptors and coregulators. Cell 108: 465–474. [DOI] [PubMed] [Google Scholar]
- Miyamoto-Sato E., Fujimori S., Ishizaka M., Hirai N., Masuoka K., et al. , 2010. A comprehensive resource of interacting protein regions for refining human transcription factor networks. PLoS ONE 5: e9289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molli P. R., Li D. Q., Murray B. W., Rayala S. K., Kumar R., 2009. PAK signaling in oncogenesis. Oncogene 28: 2545–2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mou X., Duncan D. M., Baehrecke E. H., Duncan I., 2012. Control of target gene specificity during metamorphosis by the steroid response gene E93. Proc. Natl. Acad. Sci. USA 109: 2949–2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Malley B. W., Malovannaya A., Qin J., 2012. Minireview: nuclear receptor and coregulator proteomics—2012 and beyond. Mol. Endocrinol. 26: 1646–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palijan A., Fernandes I., Verway M., Kourelis M., Bastien Y., et al. , 2009. Ligand-dependent corepressor LCoR is an attenuator of progesterone-regulated gene expression. J. Biol. Chem. 284: 30275–30287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl M. W., Horgan G. W., Dempfle L., 2002. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 30: e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddiford L., 1993. Hormones and Drosophila development, pp. 899–939 in The Development of Drosophila melanogaster, edited by Bate M., Martinez-Arias A. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Rosenfeld M. G., 2006. Sensors and signals: a coactivator/corepressor/epigenetic code for integrating signal-dependent programs of transcriptional response. Genes Dev. 20: 1405–1428. [DOI] [PubMed] [Google Scholar]
- Sapiro A. L., Ihry R. J., Buhr D. L., Konieczko K. M., Ives S. M., et al. , 2013. Rapid recombination mapping for high-throughput genetic screens in Drosophila. G3 3: 2313–2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott R. C., Juhász G., Neufeld T. P., 2007. Direct induction of autophagy by Atg1 inhibits cell growth and induces apoptotic cell death. Curr. Biol. 17: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siu M. K. Y., Chan H. Y., Kong D. S. H., Wong E. S. Y., Wong O. G. W., et al. , 2010. p21-activated kinase 4 regulates ovarian cancer cell proliferation, migration, and invasion and contributes to poor prognosis in patients. Proc. Natl. Acad. Sci. USA 107: 18622–18627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thummel C. S., 1996. Flies on steroids—Drosophila metamorphosis and the mechanisms of steroid hormone action. Trends Genet. 12: 306–310. [DOI] [PubMed] [Google Scholar]
- Thummel C. S., Burtis K. C., Hogness D. S., 1990. Spatial and temporal patterns of E74 transcription during Drosophila development. Cell 61: 101–111. [DOI] [PubMed] [Google Scholar]
- Urbanucci A., Waltering K. K., Suikki H. E., Helenius M. A., Visakorpi T., 2008. Androgen regulation of the androgen receptor coregulators. BMC Cancer 8: 219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Evans J., Andrews H. K., Beckstead R. B., Thummel C. S., et al. , 2008. A genetic screen identifies new regulators of steroid-triggered programmed cell death in Drosophila. Genetics 180: 269–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Lam G., Thummel C. S., 2010. Med24 and Mdh2 are required for Drosophila larval salivary gland cell death. Dev. Dyn. 239: 954–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Sharma D., Ren Y., Fondell J. D., 2002. A coregulatory role for the TRAP-mediator complex in androgen receptor-mediated gene expression. J. Biol. Chem. 277: 42852–42858. [DOI] [PubMed] [Google Scholar]
- Ward R. E., Reid P., Bashirullah A., D’Avino P. P., Thummel C. S., 2003. GFP in living animals reveals dynamic developmental responses to ecdysone during Drosophila metamorphosis. Dev. Biol. 256: 389–402. [DOI] [PubMed] [Google Scholar]
- Weaver T. A., White R. A., 1995. headcase, an imaginal specific gene required for adult morphogenesis in Drosophila melanogaster. Development 121: 4149–4160. [DOI] [PubMed] [Google Scholar]
- White J. H., Fernandes I., Mader S., Yang X.-J., 2004. Corepressor recruitment by agonist-bound nuclear receptors. Vitam. Horm. 68: 123–143. [DOI] [PubMed] [Google Scholar]
- White K., Tahaoglu E., Steller H., 1996. Cell killing by the Drosophila gene reaper. Science 271: 805–807. [DOI] [PubMed] [Google Scholar]
- Yang F., Li X., Sharma M., Zarnegar M., Lim B., et al. , 2001. Androgen receptor specifically interacts with a novel p21-activated kinase, PAK6. J. Biol. Chem. 276: 15345–15353. [DOI] [PubMed] [Google Scholar]
- Yin V. P., Thummel C. S., 2004. A balance between the diap1 death inhibitor and reaper and hid death inducers controls steroid-triggered cell death in Drosophila. Proc. Natl. Acad. Sci. USA 101: 8022–8027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Fondell J. D., 1999. Identification of mouse TRAP100: a transcriptional coregulatory factor for thyroid hormone and vitamin D receptors. Mol. Endocrinol. 13: 1130–1140. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.