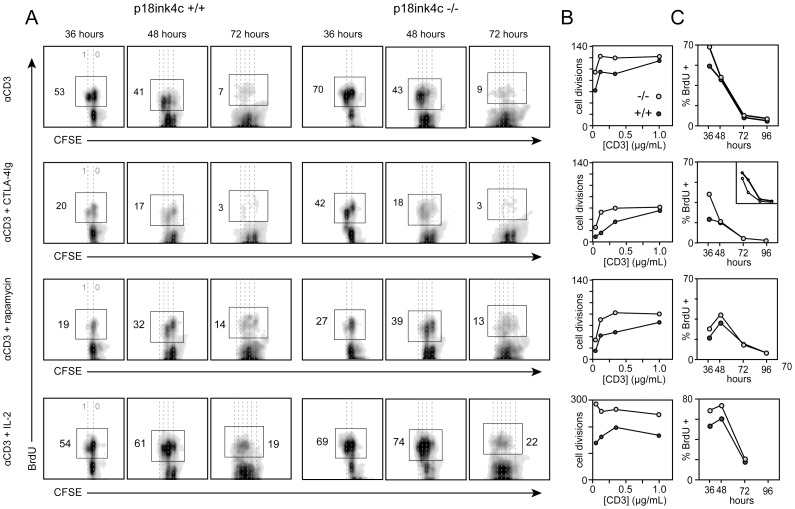
Figure 1. Kinetic analysis of DNA synthesis and cell division at the single-cell level in WT and p18ink4c-deficient CD4+ T cells.
CFSE-labeled spleen and LN cells from p18ink4c+/+ (first three columns in A, dark gray circles in B and C) or p18ink4c−/− (last three columns in A, light gray circles in B and C) mice were stimulated in vitro with soluble anti-CD3 alone (top row), or in combination with CTLA-4Ig (5 µg/mL, second row), or rapamycin (10 ng/mL, third row), or IL-2 (20 U/mL, bottom row). Cultures were subjected to 6-hour pulses of BrdU at 30, 42, 66, or 90 hours, and harvested at 36, 48, 72 or 96 hours, respectively. Non-BrdU-pulsed cultures were used to set the BrdU-positive gates at each timepoint. Dotted lines in A indicate discrete cell divisions, with undivided cells on the far right. Cumulative cell division by the CD4+ T cell subset in B was quantified as described previously (34), and C depicts the frequency of BrdU-positive cells within the CD4+ subset at each time point. The results are representative of 2–3 independent experiments.