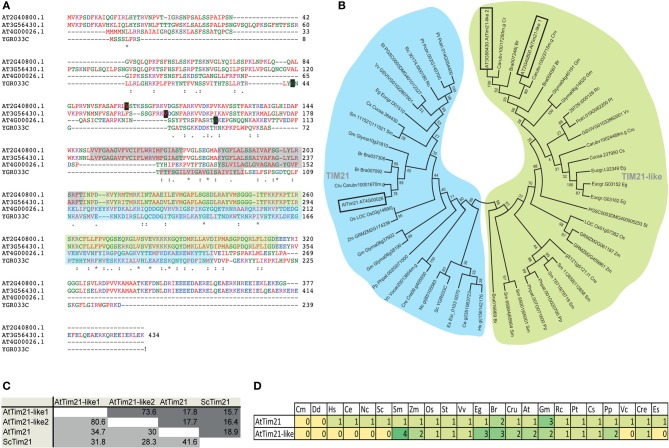
Figure 1.
Bioinformatic analysis of Tim21 and Tim21-like proteins from Arabidopsis. (A) Protein alignments of Tim21 from yeast (Saccharomyces cerevisiae, YGR033C), AtTim21 (At4g00026) and AtTim21-like 1 and 2 proteins from Arabidopsis (At2g40800 and At3g56430). The predicted mitochondrial processing sites are boxed in black and the predicted transmembrane domains are shaded in gray. The conserved Tim21 domain is shaded in blue and green. (B) Protein similarity and identity scores of ScTim21, AtTim21, and AtTim21-like 1 and 2. Similarity scores are shaded in light gray and identity scores are shaded in dark gray. (C) Phylogenetic analysis of all Tim21 and Tim21-like proteins from 24 species, the Tim21 clade is shaded in blue, and the Tim21-like clade is shaded in green. (D) The number of genes encoding Tim21 and Tim21-like proteins from each species. Abbreviations: Cm, Cyanidioschyzon merolae; Dd, Dictyostelium discoideum; Hs, Homo sapiens; Ce, Caenorhabditis elegans; Sc, Saccharomyces cerevisiae; Sm, Selaginella moellendorffii; Zm, Zea mays; Os, Oryza sativa; St, Solanum tuberosum; Vv, Vitis vinifera; Eg, Eucalyptus grandis; Br, Brassica rapa; Cru, Capsella rubella; At, Arabidopsis thaliana; Gm, Glycine max; Rc, Ricinus communis; Pt, Populus trichocarpa; Cs, Cucumis sativus; Pp, Physcomitrella patens; Vc, Volvox carteri; Cre, Chlamydomonas reinhardtii; Es, Ectocarpus siliculosus. An * (asterisk) indicates positions which have a single, fully conserved residue. A : (Colon) indicates conservation between groups of strongly similar properties-scoring > 0.5 in the Gonnet PAM 250 matrix. A . (Period) indicates conservation between groups of weakly similar properties-scoring ≤ 0.5 in the Gonnet PAM 250 matrix.