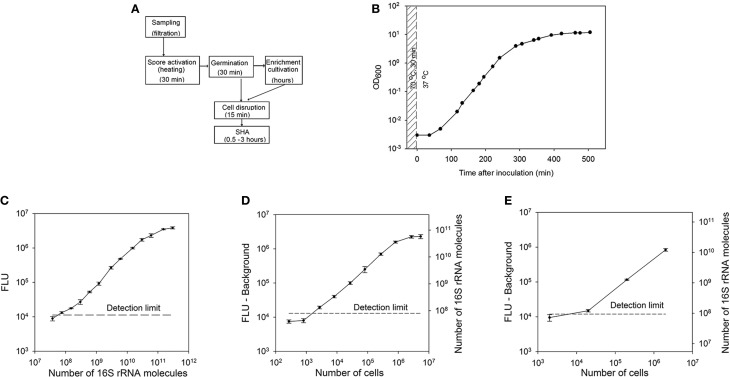
Figure 1.
Spore detection using a sandwich hybridization assay. (A) Principle of airborne spore detection using a sandwich hybridization assay. Spores collected from air are activated and cultivated in a germination medium. The first sample for analysis can be collected after approximately 30 min of germination. If the amount of spores is too low to be detected at this step, the incubation can be continued to allow a multiplication of the cells. (B) A Growth curve of B. subtilis cells. After heat activation (70°C for 30 min) B. subtilis spores were inoculated into a germination medium to a final concentration of 106 spores/ml. (C) Standard curve for the B. subtilis 16S rRNA sandwich hybridization assay. (D) SHA dilution curve for B. subtilis 16S rRNA. The dilutions of the crude extracts of B. subtilis cells collected at the exponential growth phase (OD600 = 1.4) were used as a target. The corresponding numbers of 16S rRNA molecules were calculated according to the standard curve of in vitro transcribed 16SrRNA at 260 nm. (E) 16S rRNA measured with SHA in B. subtilis spores after their activation and germination for 30 min. Corresponding numbers of 16S rRNA molecules were calculated according to the standard curve (Figure 1C). The error bars show the ±SD of three parallel experiments and the detection limit is shown as a dashed horizontal line.