Figure 5. Homology modeling of a putative DUSP5P1 derived polypeptide.
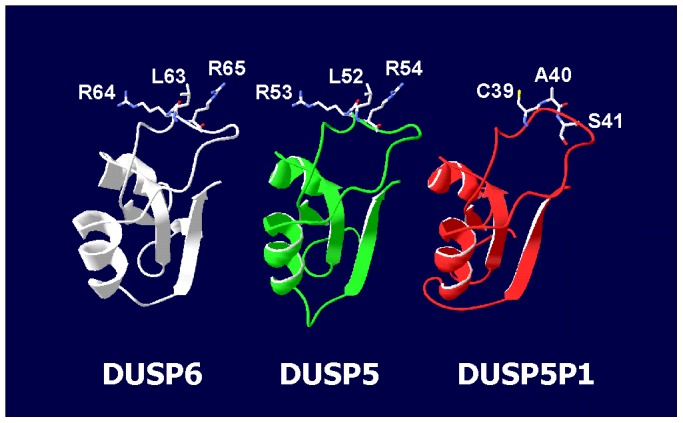
Presented is the result from an in silico homology modeling experiment using the substrate binding domain from DUSP6 [29] as template. The putative DUSP5P1 peptide (red) was predicted on the basis of a promoter scan followed by translation of all possible reading frames. This peptide (MLRKEAAAGW MVLGCRPYLA FTALSVPGSL NINLYSLVCA SPGRLWGQRA TCCQMPRSTL LLQEGSILAA VMVLN) is derived from the first open reading frame after the predicted transcription start site. In addition, the structure of the homologue region of DUSP5 (green) was predicted by using DUSP6 (white) as template. For better visibility, only the sequences from DUSP6 and DUSP5 corresponding to the predicted DUSP5P1 peptide are shown. Amino acids important for substrate binding by DUSP6 and the corresponding amino acids from DUSP5 and DUSP5P1 are highlighted.
