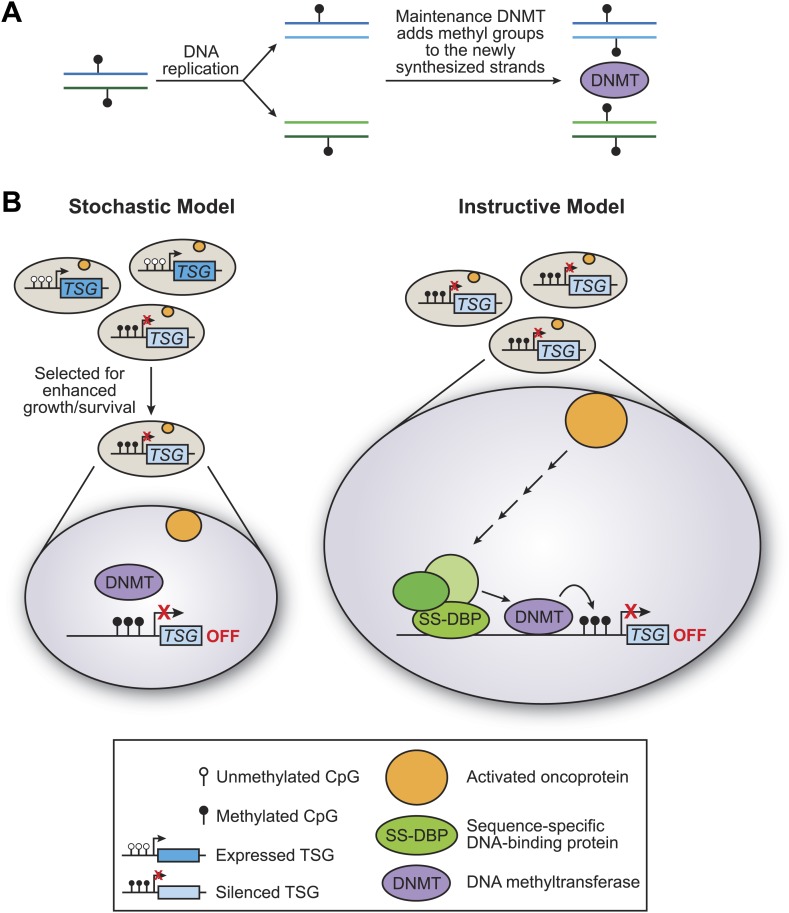
Figure 1. The stochastic and instructive models for DNA methylation of tumour suppressor genes.
(A) Prior to replication, both strands of the DNA are methylated at CpG sites. Upon DNA replication, the parental strands remain methylated, but the newly synthesized strands are not. DNA methylases (DNMT) add methyl groups to these hemi-methylated CpG sites, thereby maintaining the methylation pattern at this location in the next generation. (B) In the stochastic model (left), a cell containing methylated CpG sites and a silenced tumour suppressor gene (TSG) occurs by chance and is selected for enhanced growth/survival. Methylation of CpG residues is maintained by methylases as described in (A). In the instructive model (right), an activated oncoprotein (yellow circle) directs DNA methylation of the tumour suppressor gene via a classical transcriptional pathway involving a sequence-specific DNA binding protein (SS-DBP), a co–repressor complex (green circles) and DNA methylase. The loss of any component in this pathway results in the loss of DNA methylation and the increased expression of the tumour suppressor gene. The results of Serra et al. support the instructive model: these experiments also show that the pathway starts with an activated KRAS enzyme (shown here as a yellow circle) stimulating the expression of a kinase called PRKD1 and its substrate USP28 deubiquitinase (not shown). Phosphorylated USP28 removes a ubiquitin group from a transcription factor called ZNF304, thus increasing its concentration in the nucleus. This transcription factor (shown here as SS-DBP) binds to a specific region of DNA and recruits a scaffold protein called KAP1, an enzyme called SETDB1 that methylates histones, and a DNA methylase called DNMT1.