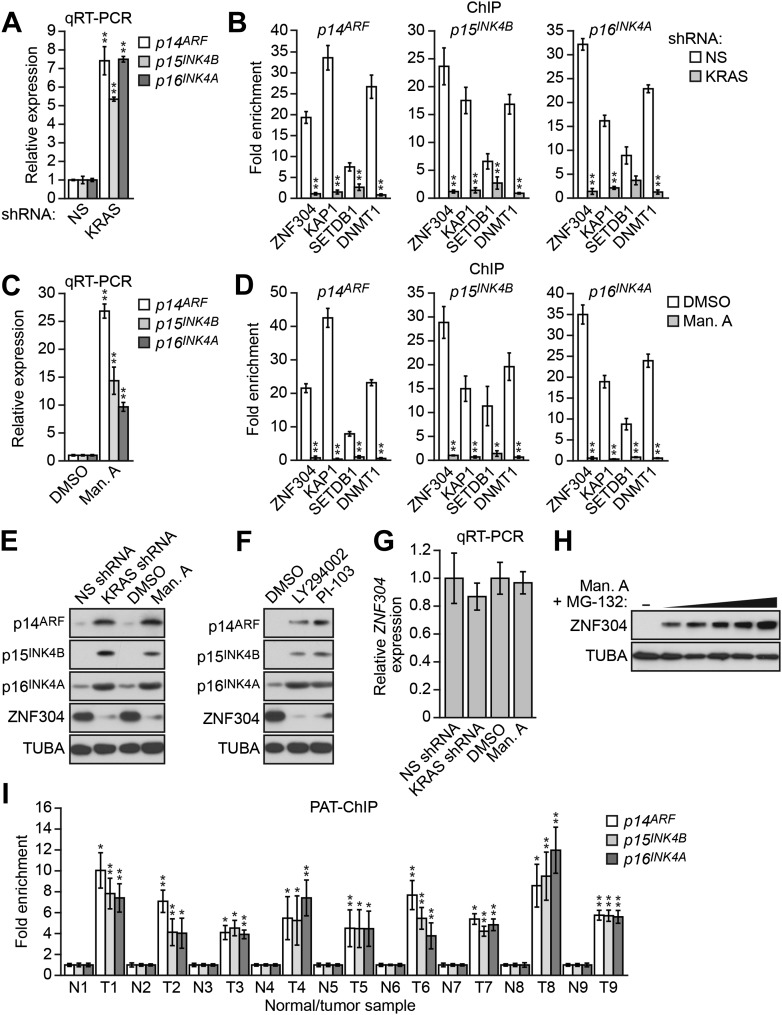
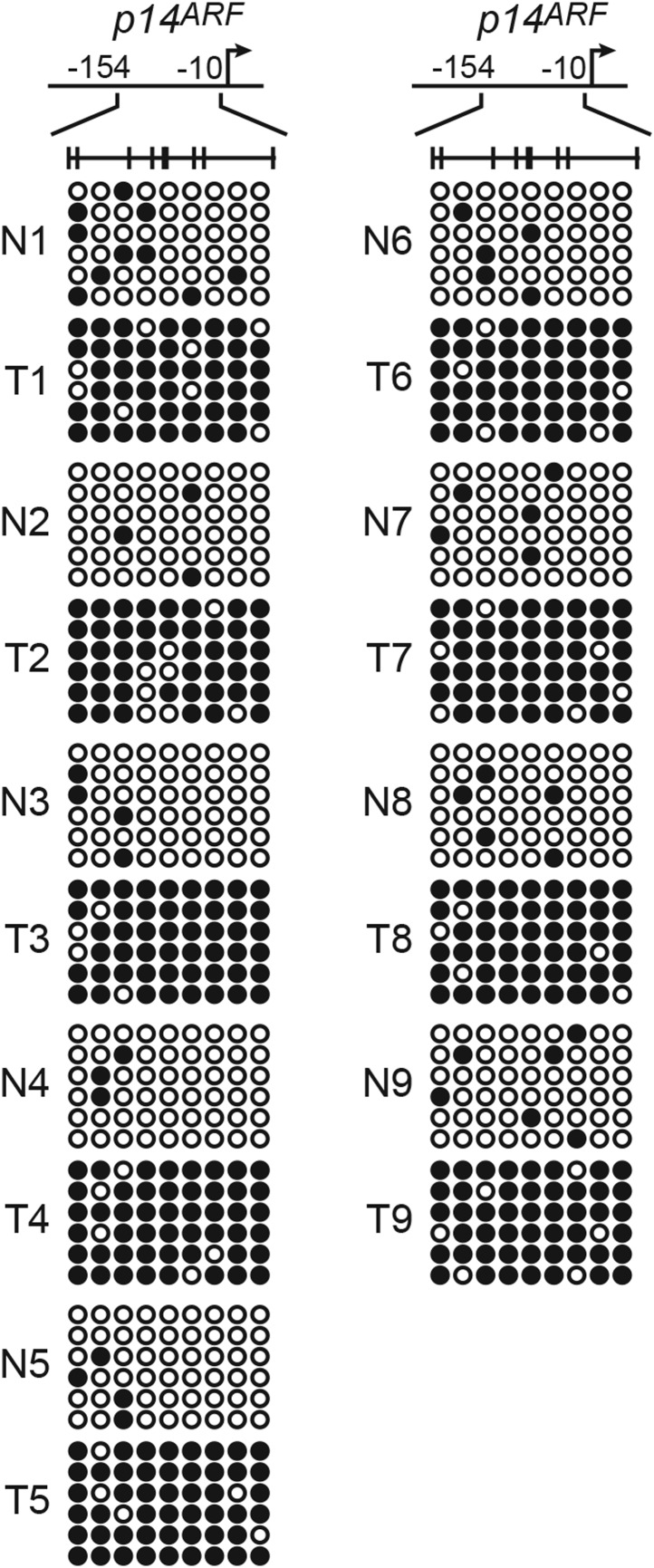
Figure 3. Activated KRAS-mediated upregulation of ZNF304 is required for transcriptional silencing of INK4-ARF.
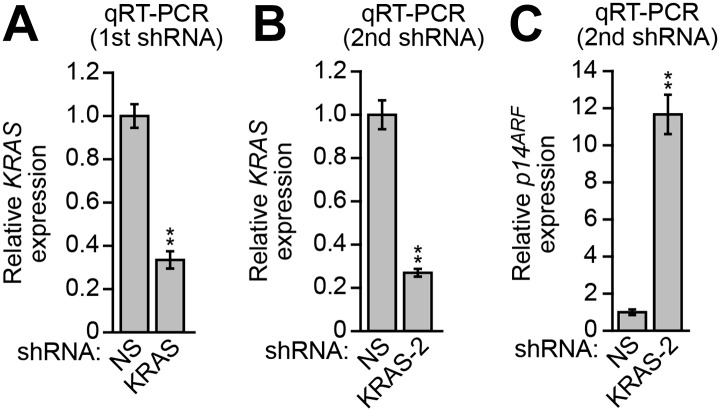
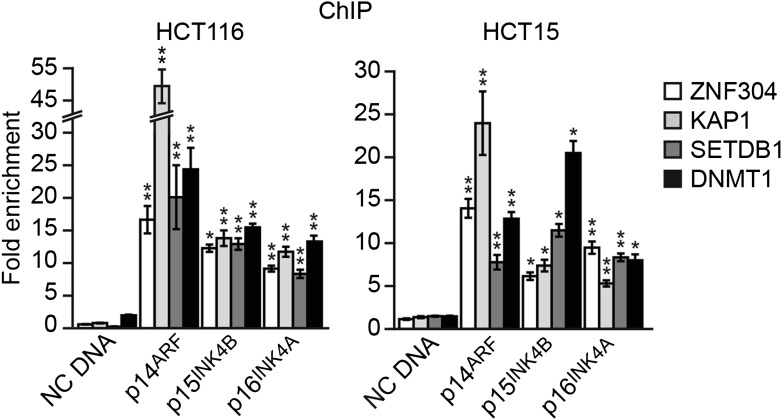
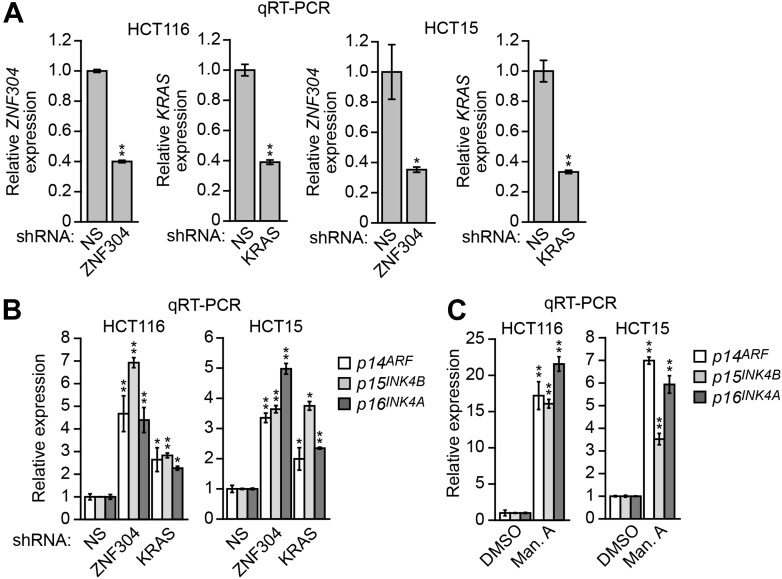
(A) qRT-PCR analysis monitoring INK4A-ARF expression in DLD-1 cells expressing a NS or KRAS shRNA. (B) ChIP analysis monitoring binding of ZNF304, KAP1, SETDB1, and DNMT1 to INK4-ARF promoters in DLD-1 cells expressing a NS or KRAS shRNA. (C) qRT-PCR analysis monitoring INK4A-ARF expression in DLD-1 cells treated with DMSO or manumycin A (Man. A). The results were normalized to DMSO, which was set to 1. (D) ChIP analysis monitoring binding of ZNF304, KAP1, SETDB1, and DNMT1 to INK4-ARF promoters in DLD-1 cells treated with DMSO or Man. A. (E) Immunoblot analysis showing INK4-ARF levels in DLD-1 cells treated with a NS or KRAS shRNA, or DMSO or Man. A. (F) Immunoblot analysis showing INK4-ARF levels in DLD-1 cells treated with DMSO, LY294002, or PI-103. (G) qRT-PCR analysis monitoring ZNF304 expression in DLD-1 cells treated with a NS or KRAS shRNA, or DMSO or Man. A. (H) Immunoblot analysis showing ZNF304 levels in DLD-1 cells treated with Man. A for 24 hr and 0–10 µM MG-132 for 4 hr. (I) PAT-ChIP analysis monitoring binding of ZNF304 to INK4-ARF promoters in matched adjacent normal (N) and KRAS-positive CRC human tumor (T) samples. Results were normalized to normal samples, which were set to 1. Data are represented as mean ± SD. *p≤0.05, **p≤0.01. Results from experiments validating KRAS knockdown efficiency and the role of KRAS in repressing p14ARF expression, as well as experiments validating the role of ZNF304 and its corepressors in INK4-ARF silencing in other KRAS-positive CRC cell lines, are presented in Figure 3—figure supplements 1–4.