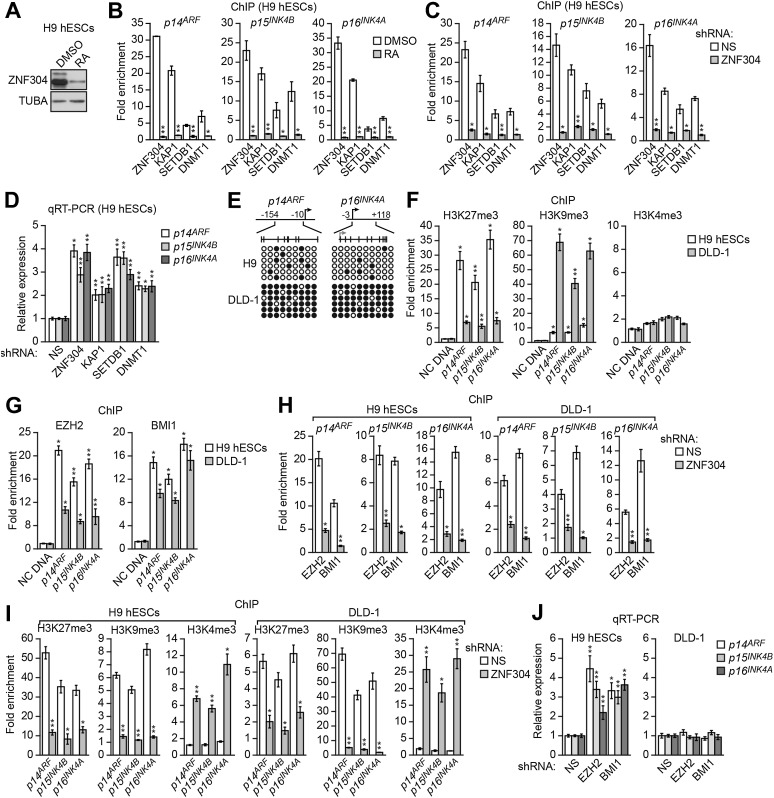
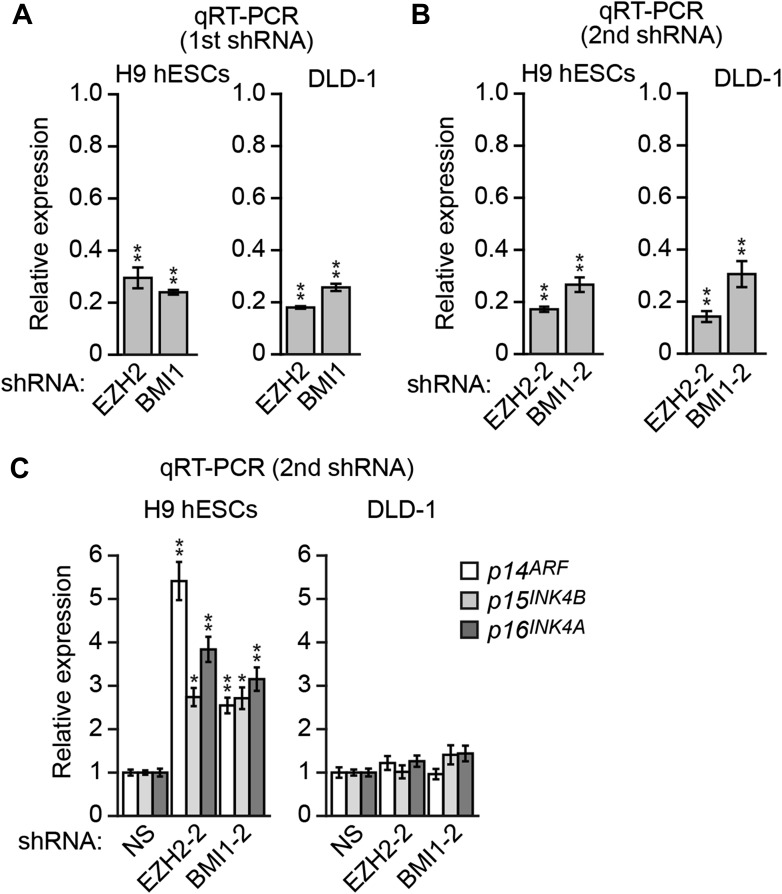
Figure 7. ZNF304 also directs transcriptional silencing of INK4-ARF in hESCs.
(A) Immunoblot analysis showing ZNF304 levels in undifferentiated (DMSO) or retinoic acid (RA)-treated hESCs. (B and C) ChIP analysis monitoring binding of ZNF304, KAP1, SETDB1, and DNMT1 to INK4-ARF in undifferentiated or RA-treated hESCs (B) or in hESCs expressing a NS or ZNF304 shRNA (C). (D) qRT-PCR analysis monitoring INK4-ARF expression in hESCs expressing a NS, ZNF304, KAP1, SETDB1, or DNMT1 shRNA. (E) Bisulfite sequencing analysis of the p14ARF and p16INK4A promoters in H9 hESCs and DLD-1 cells. (F and G) ChIP analysis monitoring enrichment of H3K27me3, H3K9me3, and H3K4me3 (F) and EZH2 and BMI1 (G) at INK4-ARF or an irrelevant DNA region (NC) in H9 hESCs and DLD-1 cells. (H and I) ChIP analysis monitoring binding of EZH2 and BMI1 (H) and H3K27me3, H3K9me3 and H3K4me3 (I) at INK4-ARF in H9 hESCs and DLD-1 cells expressing a NS or ZNF304 shRNA. (J) qRT-PCR analysis monitoring INK4-ARF expression in H9 hESCs and DLD-1 cells expressing an NS, EZH2, or BMI1 shRNA. Data are represented as mean ± SD. *p≤0.05, **p≤0.01. Control experiments related to Figure 7 are shown in Figure 7—figure supplements 1,2.
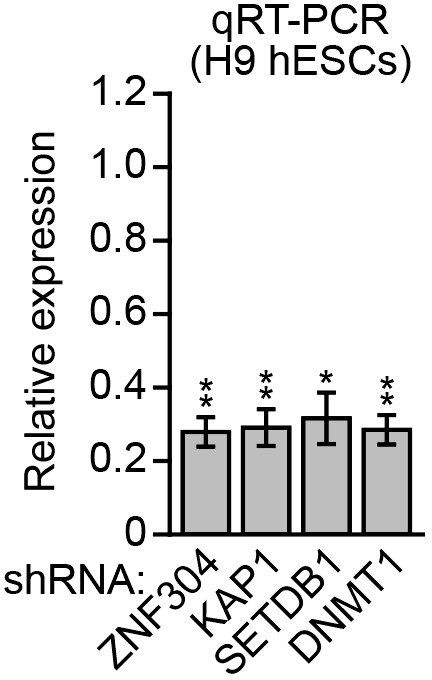
Figure 7—figure supplement 1. Knockdown efficiencies in H9 hESCs.