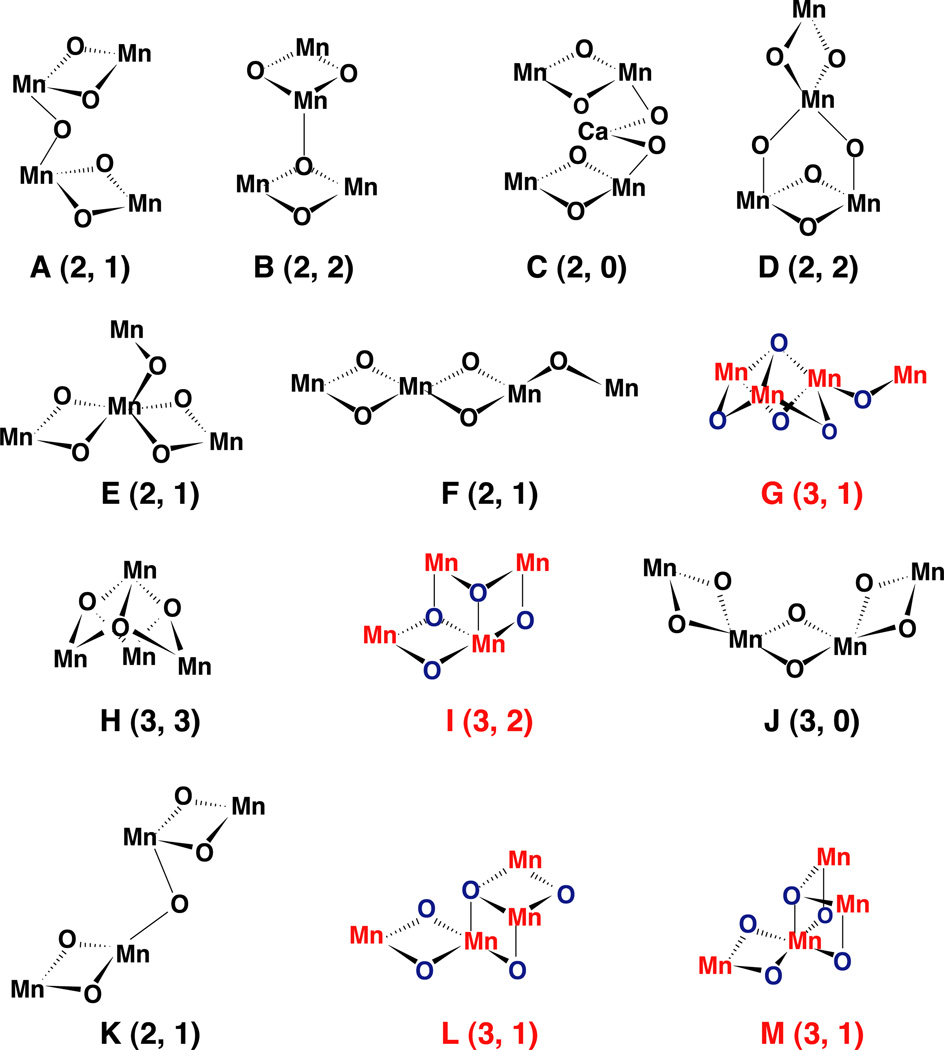
Fig. 2.
Topological models for the Mn complex identified as compatible with the EXAFS data by DeRose et al. [1] and Robblee at al. [31]. The nomenclature is identical to the one we have used in our two earlier publications by DeRose et al. and Robblee et al. The numbers in parenthesis are the number of short 2.7- to 2.8-Å Mn–Mn vectors and long 3.3- to 3.4-Å Mn–Mn vectors. The topologies in color (options G, I, L and M) all have three short 2.7- to 2.8-Å Mn–Mn vectors and are in better agreement with the X-ray diffraction studies; options E, F and G have been identified as compatible with EPR and ENDOR data. Option G or a topological isomer seems to be the best model by all presently available criteria (X-ray diffraction, EXAFS and EPR).