Abstract
AIM
To investigate the protective effects of the natural medicinal monomer ecdysterone (ECR) with estrogenic activity against oxidative damage in human lens epithelial cells B3 (HLE-B3) caused by hydrogen peroxide 21(H2O2) and to pursue the possible mitochondrial proteomic regularity of the protective effects.
METHODS
HLE-B3 cells were treated with H2O2 (300µmol/L), β-estuarial (E2; 10−8mol/L) and H2O2, ECR (10−6mol/L) and H2O2, or left untreated. Altered expression of all mitochondrial proteins was analyzed by protein array and surface-enhanced laser desorption ionization time of flight mass spectrometry (SELDI-TOF-MS). The mass/charge (M/Z) ratios of each peak were tested by the Kruskal-Wallis rank sum test, and the protein peak value of the M/Z ratio for each treatment by pair comparison was analyzed with the Nemenyi test.
RESULTS
H2O2 up-regulated expression of two protein spots (with M/Z of 6 532 and 6 809). When E2 mitigated the oxidative damage, the expression of one protein spot (M/Z 6 532) was down-regulated. In contrast, ECR down-regulated both of protein spots (M/Z 6 532 and 6 809).
CONCLUSION
ECR could effectively inhibite H2O2 induced oxidative damage in HLE-B3 cells. The protein spot at M/Z of 6 532 might be the target spot of ECR against oxidative damage induced by H2O2.
Keywords: ecdysterone, mitochondrial proteomics, lens epithelial cell, senile cataract
INTRODUCTION
Epidemiology data indicated the morbility of senile cataract in woman is higher than that in man; especially obvious in the post-menopause woman[1]-[2]. Clinical and fundamental studies find the development of senile cataract is relative about deficiency of estrogen[3], the oxidative damage extenuated by estrogen in the lens, inhibiting apoptosis in the lens epithelia cells (LEC), and playing role in the protection of cataract[4]-[8]. Current studies demonstrated that Chinese drug abundant in phytoestrogen had estrogen-like activity, such as achyranthis, psoralea fruits, kudzuvine root, epimedium, black cohosh root, giant knotweed rhizome, which were not succedaneous in protection of cancer and cerebrovascular disease[9]. Ecdysterone (ECR), an effective monomer in achyranthis, could enhance the expression of estrogen receptor (ER) in tissue, accentuate the effect of endogenous estrogen and has estrogen-like activity[10].
Previous works found that ECR increased expression of ER β and activities of superoxide dismutase (SOD), glutathione peroxidase (GSH-px) and catalase (CAT) on human lens epithelial cell B3 (HLE-B3). It indicated that ECR had estrogen-like activity and protected HLE-B3 against oxidative damage[11]-[13]. Our study, based on the foundation of protective effects of ecdysterone on oxidative damage of HLE-B3, the change of expression of all mitochondrial proteins in HLE-B3 after oxidative damage was assayed and analyzed by protein array and surface-enhanced laser desorption ionization time of flight mass spectrometry (SELDI-TOF-MS) proteomics technology to seek differential proteins.
MATERIALS AND METHODS
Materials
Reagents and apparatus
HLE-B3 cells were provided by the Research Institute of Ophthalmology of Guangzhou Zhongshan University. The following reagents were purchased: E2 (Sigma, USA;97%), ecdysterone (ECR) (National institute for the control of pharmaceutical and biological products, China, 96.3%), 30% H2O2 (Shanghai reagent plant, China), dulbecco's modified eagle's medium (DMEM; GIBCO, USA), tryptase (Amresco, USA), 3-[(3-cholamidopropyl) dimethylammonio] propanesulfonic acid (CHAPS), ethylenediaminotetraacetic acid (EDTA; Augus, USA); trifluoro-acetic acid (TFA), urea, and dithiothreitol, (DTT; Sigma). The Mitochondria Isolation Kit for Cultured Cells was purchased from Pierce (USA) and the Mitochondria Protein Isolation Kit from Xinghan (China). The ELISA reading instrument was a BioTek ELX808 (USA) product. The PBS II+ type SELDI-TOF-MS and 96-hole Bio-processor protein chip work platform were both from Ciphergen Biosystem Inc (USA). The Biophotometer 6131 protein nucleic acid radiometer was from Eppendorf (Germany).
Methods
HLE-B3 culture and generation
The frozen cells were thawed in a 37°C water bath. The cell suspension was diluted 10 times in culture medium, centrifuged, and then washed three times. The cells were inoculated in a culture flask at 5×105 cells/mL, and grown at 37°C with 5% CO2. Every 2-3d, cells were subcultured.
Detection of mitochondrial proteome alteration in HLE-B3 with SELDI-TOF-MS proteinchip array
Optimal drug concentrations were obtained by the MTT assay[11]. HLE-B3 cells were treated with 300µmol/L H2O2 alone, or in combination with 10−8mol/L E2 or 10−6mol/L ECR, or treated with culture medium (control). The DMEM culture medium was removed after 24h, and then cells were washed three times with phosphate-buffered saline (PBS). After cells were scraped into 1mL PBS and centrifuged at 700×g for 5min. the supernatant was removed and 1mL PBS was added to resuspend the cells. The cells were counted and aliquotted at 2×107 cells/mL in 2.0mL microcentrifuge tubes and centrifuged at 850×g for 2min. Again, the supernatant was removed, and 800µL mitochondrial separation reagent A was added. The solution was swirled for 5s, incubated for 2 min on ice, and then the cell suspension was transferred to a Dounce Tissue Grinder for homogenization. The lysed cells were returned to the original tube and mitochondrial separation reagent C (800µL) and mitochondrial separation reagent A (200µL) were added to the tube, which was inverted several times to mix and then centrifuged at 700×g for 10min at 4°C. The resulting supernatant was extracted and then centrifuged at 3 000×g for 15min at 4°C. The supernatant was removed, and 500µL mitochondrial separation reagent C was added. The sample was centrifuged at 12 000×g for 5min at 4°C. The supernatant was removed to get the pellet, i.e. HLE-B3 mitochondria. The mitochondrial protein concentration was detected by the Coomassie brilliant blue method as follows: 200µL mitochondrial protein extraction reagent was added, oscillated for 15min at 4°C, mixed and then centrifuged at 12 000×g for 15min at 4°C. After the supernatant was extracted, the protein concentration was determined and adjusted to 5mg/mL.
Each mitochondrial protein sample was combined with a CM10 chip, 3 spots on the chip for each treatment group, in a wet box according to the manufacturer's instructions. Binding buffer (50mmol/L sodium acetate, pH4.0; 3µL) was added to each sample spot, immersed for 3 times, 5min each time. The buffer was removed, and 5µL buffer was added immediately to dilute the protein sample to 1.8mg/mL. After 1h at room temperature, the buffer was removed and 3µL binding buffer was added to each spot, and then removed after 5min; this procedure was repeated 3 times. HPLC-grade water (3µL) was added to each spot and then removed immediately; this procedure was repeated 2 times. The chips were air-dried for 20min, and Sinapic Acid (SPA) solution was added followed by air drying once more. The SELDI-MS data were corrected to an error of less than 0.1% molecular weight using a standard molecular weight chip. The CM10 chip-combined proteins were detected by mass meter reading. The unified analysis parameter was used, with the laser intensity set at 185, detection sensitivity at 7, a detection top limit of 100 000M/Z, optimized collection data range of 2 000-20 000M/Z, and a signal collection location of 20-80. Each sample had an average of 144 spots. Initial data were exported in xlm format using Proteinchip Software 3.2 [14].
Statistical Analysis
SELDI-TOF-MS data are presented as the mean±standard error of the mean (SEM). Statistical analysis was performed with ZUCI-ProteinChip Data Analyze System software. designed by the Cancer Institute of Zhejiang University. Noise was removed from the initial data for each sample using an undecimated discrete wavelet transformation, and the baseline was subtracted. Peaks under 2 000 were discarded. Each sample peak was found by partly extremum, and peaks with a signal to noise ratio less than 4 were discarded. The quantity of peaks from each sample, and their M/Z values, were all different. Peaks whose disparities were less than 0.3% in each sample were divided into a separate category. Peaks occurring in the sample at less than 10% were excised. The intensity of found peaks in each sample was processed uniformly. Peaks were selected as a characteristic vector by statistics filter combined model dependent screening. Each peak value (M/Z ratio) was analyzed by Kruskal-Wallis rank sum test. Peaks for which the P is minimum were chosen for further analysis. Pairwise comparison of protein peak value for each defined M/C ratio in different treatment groups were assessed by the Nemenyi test. Differences were considered statistically significant when the P was less than 0.05[14].
RESULTS
Mitochondrial Proteome of HLE-B3 induced by H2O2
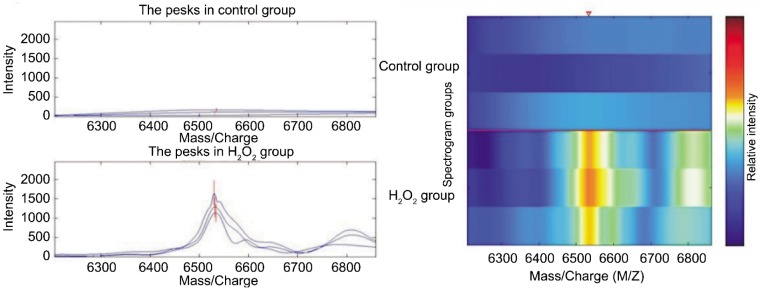
There were 50 protein spots in CM10 chip binded with HLE-B3 induced by H2O2, The expression was up-regulated in those two protein spots, the M/Z ratios of which were 6 532 and 6 809. The values of those two sample were 13.6 and 4.8 times higher than those of the control respectively (Table 1 and Figure 1). There were significant difference between H2O2 group and control group (P<0.05).
Table 1. Mitochondrial proteomics of HLE-B3 Induced by H2O2.
| M/Z | Average intensity |
Multiples of the intensity change | |
| Control group | H2O2 group | ||
| 6532 | 107.76±70.84 | 1460.72±267.57a | +13.6 |
| 6809 | 109.59±31.19 | 522.09±200.20a | +4.8 |
aP<0.05, compared with control group.
(n=3, x±s)
Figure 1. Comparison of expressions of CM10 chip mitochondrial proteome between H2O2 group and control group.
Mitochondrial Proteome of Protective Effects of E2 on Oxidative Damage in HLE-B3
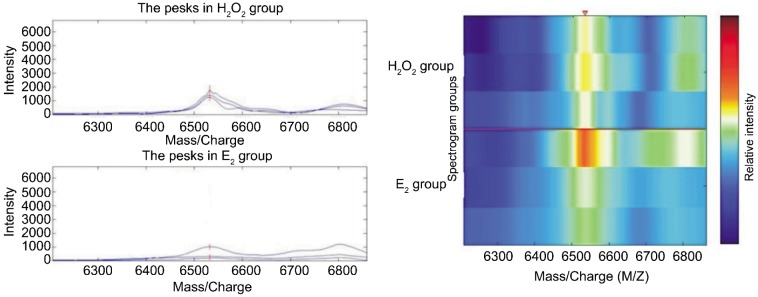
There were 46 protein spots in CM10 chip binded with HLE-B3 after protective effects of E2 on oxidative damage induced by H2O2. The expression was down-regulated in protein spot with a M/Z of 6 532. It was 2.8 times lower than that of H2O2 group (Table 2, Figure 2). Difference was significant between E2 group and H2O2 group (P<0.05).
Table 2. Mitochondrial proteome of protective effects of E2 on oxidative damage in HLE-B3.
| M/Z | Average intensity |
Multiples of the intensity change | |
| H2O2 group | E2 group | ||
| 6532 | 1460.72±267.57 | 519.59±138.72a | -2.8 |
aP<0.05, compared with H2O2 group.
(n=3, x±s)
Figure 2. Comparison of expressions of CM10 chip mitochondrial proteome between E2 group and H2O2 group.
Mitochondrial Proteomics of Protective Effects of ECR on Oxidative Damage in HLE-B3
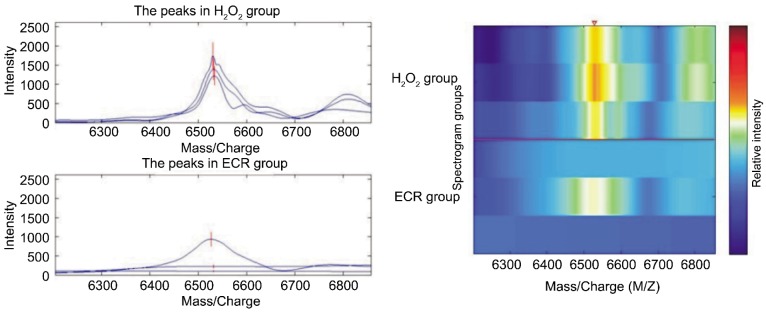
There were 49 protein spots in CM10 chip binded with HLE-B3 after protective effects of ECR on oxidative damage induced by H2O2. The expression was obviously down-regulated in those protein spots with the M/Z ratios of 6 532 and 6 809. They were 3.4 and 2.6 times lower than those of H2O2 group (Table 3, Figure 3). Difference was significant between ECR group and H2O2 group (P<0.05).
Table 3. Mitochondrial Proteomics of Protective Effects of ECR on Oxidative damage of HLE-B3.
| M/Z | Average intensity |
Multiples of the intensity change | |
| H2O2 group | ECR group | ||
| 6 532 | 1460.72±267.57 | 420.22±449.56a | -3.4 |
| 6 809 | 522.09±200.20 | 197.29±90.35a | -2.6 |
aP<0.05, compared with H2O2 group.
(n=3, x±s)
Figure 3. Comparison of expressions of CM10 chip mitochondrial proteome between ECR group and H2O2 group.
As shown in all the above tables and figures, there was a protein spot with a M/Z of 6 502 in ECR group, E2 group, H2O2 group and control group. Its expression of H2O2 group (1 460.72±267.57) was significantly higher than that of control group (107.76±70.84) (P<0.05). The expressions in ECR group (420.22±449.56) and E2 group (519.59±138.72) were all obviously lower than H2O2 group (P<0.05). H2O2 up-regulated expression of two protein spots (with M/Z of 6 532 and 6 809). When E2 mitigated the oxidative damage, the expression of one protein spot (M/Z 6 532) was down-regulated. In contrast, ECR down-regulated both of protein spots (M/Z 6 532 and 6 809).
DISCUSSION
Mitochondria are essential for cell's life and death. Mitochondrial proteomics, at subcellular level, mainly identifies the total protein in mitochondria. The research of mitochondrial proteomics could comprehend its structure and function, find the effect of mitochondrial proteomics in generation and development of disease, and provide clue to seek especial protein which is intimately related to disease. By comparing different research proteomic techniques, we found different mitochondrial proteins in pathological conditions and provided important target for diagnosis and treatment. Mitochondrial proteomic analysis has been performed in cardiac failure, liver cancer, and amyotrophic lateral sclerosis, among others. However, there are only two reports about eye diseases. Nordgaard et al[15] demonstrated that age-related macular degeneration (AMD) induces differential expression of eight different spots. These results are consistent with the hypothesis that mitochondrial dysfunction is associated with AMD, and suggest specific pathophysiological mechanisms involving altered mitochondrial translation, import of nuclear-encoded proteins, and adenosine triphosphate (ATP) synthase activity. Saraswathy and Rao [16] found alterations in retinal mitochondrial protein levels in response to oxidative stress during the early phase of experimental autoimmune uveitis (EAU). The presence of mitochondrial-specific oxidative stress-related proteins in the early EAU retina, along with the down-regulation of ATP synthase, provides early evidence of stress-related retinal damage. The presence of high levels of alphaA and betaB2 crystallin in the mitochondria may prevent cell death during early EAU. Neither of these addresses the mitochondrial proteomics of lens epithelial cells forming cataracts. There was no report about mitochondrial proteomics of lens epithelial cell and cataract
SELDI-TOF-MS ProteinChip array which is a new research method of proteomics consist of protein separation and purification and mass spectrometric detection. Though different protein chip modification, it selectively captures different protein in sample, consumedly decreases protein complexity in sample. It maintains analyzing diversiform sample diversiform protein, puts every protein to a cross-reference value background. It directly detects relatively initial biological specimen and discovers many low concentration, low molecular weight protein which is masked. This technique, which could detect small protein whose concentration is 10−15moL (fmol) and obtain data of hundreds of protein in sample, is a useful tool to analyze cell proteome. And it has many advantages, such as high resolution and repeatability, simple application and high sensitivity. So, its application in proteomics becomes more and more wide. Yet there has been no report on mitochondrial proteomics of lens epithelium cells using this technique so far.
Previous works showed that H2O2 induced oxidative damage in HLE-B3. E2 and ECR increased HLE-B3 proliferation, as well as enhancing SOD, GSH-px and CAT activities and decreasing malondialdehyde (MDA) expression to prevent oxidative damage. Both of E2 and ECR also increased DNA content in the HLE-B3 nucleus, and stabilized the mitochondrial transmembrane potential to prevent apoptosis. Increased ERβ expression reduced oxidative damage in HLE-B3 cells. These results demonstrated that ECR might have estrogen-like activity, increases antioxidant activity in HLE-B3 cells, decrease apoptosis, and protect HLE-B3 cells against oxidative damage[11]-[13].
According to other researches, this study applied three chip, including H4 chip, WCX2 chip and CM10 chip which were combined with proteins in sample respectively[17],[18]. We found that protein spots combined with CM10 chip were the most of all chips, and protein spots combined with CM10 chip almost included all protein spots combined with H4 chip and WCX2 chip. Estrogen reduces oxidative damage and inhibits the LEC apoptosis involved in cataract development. Fifty protein spots were detected in the samples of H2O2 group, 46 protein spots in those of E2 group and 49 protein spots of ECR group. Differential expression occurred in two spots (M/Z 6 532 and 6 809). After H2O2-induced oxidative damage, the expression of these two protein spots was up-regulated compared to untreated cells. E2 down-regulated expression of the spot with the M/Z ratio of 6 532, while both were down-regulated by ECR. The protein spot with an M/Z ratio of 6 532 may be an important target of ECR, to protect HLE-B3 cells against oxidative damage induced by H2O2.
Next we identified the mitochondrial proteins to elucidate the pathogenesis of senile cataract. According to the M/Z ratio combined with the isoelectric point range from chip, we retrieved matched protein information in the Swiss-Prot data bank. There were seven proteins that matched the protein spot with an M/Z ratio of 6 532: putative protein FAM41C, 40S ribosomal protein S29, β-alexin 136, surface active agent associated protein 2, putative uncharacterized protein C10orf99, uncharacterized protein C21orf119 and putative antigen processing associated transporter-2 connected polypeptide. Which associated with HLE-B3 apoptosis, cells from oxidative damage and senile cataract -related protein is only one, namely the 40S ribosomal protein S29 (ribosomal protein S29, RPS29). The protein may play important role in protecting ECR against oxidative damage in HLE-B3 induced by H2O2. It may be the drug target against senile cataract.
Ribosomal proteins, the major constituents of ribosomes, catalyze protein synthesis in the cytoplasm. The eukaryotic ribosome is composed of a large (60S) and a small (40S) subunit consisting of three RNAs and 46 proteins and one RNA and 33 proteins, respectively[19]. Zhang et al[20]found that numerous transcripts exhibited altered levels of gene expression. One transcript exhibiting a decreased level of expression in cataracts compared with normal lenses was identified as encoding ribosomal protein L21. Three additional ribosomal proteins, L15, L13a, and L7a, also exhibited decreased expression in cataracts compared with normal human lenses. By contrast, the levels of elongation factor (EF)-1alpha1 and eukaryotic initiation factor (eIF)-4E remained unchanged. These results provide evidence that age-related cataracts in humans are associated with decreased expression of L21 and other ribosomal proteins. Therefore, modulation of protein synthesis and/or other functions mediated by ribosomal proteins may be associated with the formation of age-related cataracts.
Mitochondrial ribosomal proteins S29 (Mitochondrial ribosomal proteins S29, MRPS29) is a mitochondrial ribosomal small subunit 40S protein component. MRPs constitute the mitochondrial translation machinery, but also has other functions. In 1996, Kondoh et al[21] first identified the ribosomal protein S29 (ribosomal protein S29, RPS29) can increase Krev-1 gene expression of the inhibitory activity of the product of the malignant proliferation of cells; Khanna et al[22] studied the apoptosis related genes found that apoptosis of thymus cells and normal thymocytes RPS29 gene expression was significantly increased; the gene transfection of human Hela cells and rat thymocytes apoptosis intensified, show that RPS29 in the occurrence of apoptosis in primary a key role. Tang et al[23] experimental study found that the RPS29 gene product can inhibit the proliferation of fibroblasts, and can promote into the occurrence of apoptosis in fibroblasts. Cavdar Koc et al[24] found that two closely related to the apoptosis protein GTP-binding protein DAP3 (death associated protein, 3) and PDCD9 (programmed cell death protein 9)are actually of mitochondrial ribosomal small subunit protein MRPS29 and MRPS30, and further clarify the mitochondrial ribosome and apoptosis are closely related. Furthermore, ECR can be reduced within HLEC 40S ribosomal protein S29, affecting protein synthesis within HLEC and inhibiting oxidative damage in HLEC. Therefore, the 40S ribosomal protein S29 may be the target of ECR protection against H2O2-induced oxidative damage.
Compared with other proteomics techniques, SELDI-TOF-MS has higher discrimination in the low molecular weight range (from 2 to 200kDa). Differentially expressed proteins, mostly low-molecular-weight proteins generated in a specific environment, usually are some metabolic products, abnormally located proteins, modified proteins, neurotransmitters, polypeptides or cytokines. Therefore, differentially expressed mitochondrial proteins detected in this study were probably not reported previously as being involved in senile cataract pathogenesis. Further identifying and studying these mitochondrial proteins could be helpful to identify their roles in senile cataracts, as well as the potential drug targets.
Acknowledgments
Foundation: Supported by Fujian Province Health Department Fund (No.2009-1-30); Fujian Province Department of Education Issues (No.JA10176)
Conflicts of Interest: Feng CY, None; Huang XR, None; Qi MX, None; Tang SW, None; Chen S, None; Hu YH, None; Ke FJ, None; Wang X, None.
REFERENCES
- 1.Klein BE, Klein R, Linton KL. Revalence of age related lens opacities in a population. The Beaver Dam Eye study. Ophthalmology. 1992;99(4):546–552. doi: 10.1016/s0161-6420(92)31934-7. [DOI] [PubMed] [Google Scholar]
- 2.Cumming RG, Mitchell P. Hormone replacement therapy, reproductive factors, and cataract. The Bluc Mountains Eye Study, Am J Epidemiol. 1997;145(3):242–249. doi: 10.1093/oxfordjournals.aje.a009097. [DOI] [PubMed] [Google Scholar]
- 3.Dolatowska E. The evaluation of estradiol and FSH serum levels in menopausal women with primary cataract. Klin Oczna. 2002;104(5–6):357–361. [PubMed] [Google Scholar]
- 4.Wang X, Simpkins JW, Dykens JA, Cammarata PR. xidative damage to human lens epithelial cells in culture: estrogen protection of mitochondrial potential, ATP, and cell viability. Invest Ophthalmol Vis Sci. 2003;44(5):2067–2075. doi: 10.1167/iovs.02-0841. [DOI] [PubMed] [Google Scholar]
- 5.Moor AN, Flynn JM, Gottipati S, Giblin FJ, Cammarata PR. 17beta-estradiol stimulates MAPK signaling pathway in human lens epithelial cell cultures preventing collapse of mitochondrial membrane potential during acute oxidative stress. Mitochondrion. 2005;5(4):235–247. doi: 10.1016/j.mito.2005.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moor AN, Gottipati S, Mallet RT, Sun J, Giblin FJ, Roque R, Cammarata PR. A putative mitochondrial mechanism for antioxidative cytoprotection by 17 beta-estradiol. Exp Eye Res. 2004;78(5):933–944. doi: 10.1016/j.exer.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 7.Flynn JM, Cammarata PR. Estradiol attenuates mitochondrial depolarization in polyol-stressed lens epithelial cells. Mol Vis. 2006;12:271–282. [PubMed] [Google Scholar]
- 8.Gottipati S, Cammarata PR. Mitochondrial super-oxide dismutase activation with 17 beta-estradiol-treated human lens epithelial cells. Mol Vis. 2008;14:898–905. [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao PW, Wang DW, Wang LQ. Expression study in screening estrogen-like ability of ten raditional Chinese drug using uterus increased weight. J Beijing University TCM. 2006;29(10):686–689. [Google Scholar]
- 10.Zhao PW, Niu Z, Wang JF. Influence of Isopsoralen and ecdysterone in proliferation of human breast carcinoma cell. J Beijing University TCM. 2007;30(4):242–245. [Google Scholar]
- 11.Qi MX, Huang XR, Zhang KL, Guo N. Isopsoralen protecting against apoptosis of human lens epithelial cell. Chinese J Clin Pharmacol Therapeutics. 2009;14(12):1371–1374. [Google Scholar]
- 12.Huang XR, Qi MX, Zhang KL, Guo N. Upregulatory effect of isopsoralen on expression of estrogen receptor in human lens epithelial cell. Chin J Pathophysi. 2010;26(9):1844–1848. [Google Scholar]
- 13.Feng CY, Huang XR, Qi MX, Tang SW, Guo N, Hu YH. The protective effects and mechanisms of Isopsoralen in oxidative damage of human lens epithelial cell. Chia J Ophthalmol. 2011;47(4):353–355. [Google Scholar]
- 14.Hu YH, Huang XR, Qi MX, Hou BY. Study on Proteomics of inhibitory effects of elemene on proliferation of human lens epithelial cell. Chia J Ophthalmol. 2010;46(5):427–431. [PubMed] [Google Scholar]
- 15.Nordgaard CL, Karunadharma PP, Feng X, Olsen TW, Ferrington DA. Mitochondrial proteomics of the retinal pigment epithelium at progressive stages of age- related macular degeneration. Invest Ophthalmol Vis Sci. 2008;49(7):2848–2855. doi: 10.1167/iovs.07-1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saraswathy S, Rao NA. Mitochondrial proteomics in experimental autoimmune uveitis oxidative stress. Invest Ophthalmol Vis Sci. 2009;50(12):5559–5566. doi: 10.1167/iovs.08-2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Q, Liu Y, Ren YF. Different expressed protein was detected using SELDI-TOF-MS. Applied Journal of General Practice. 2008;6(4):333–334. [Google Scholar]
- 18.Ge Z, Zhu YL, Zhong X, Yu JK, Zheng S. The influence of Helicobacter pylori associated protein A in human gastric adenocarcinoma epithelium cell proteome. Cell Biology. 2006;28(4):603–610. [Google Scholar]
- 19.Wool IG, Chan YL, Gluck A. Structure and evolution of mammalian ribosomal proteins. Biochem Cell Biol. 1995;73(11–12):933–947. doi: 10.1139/o95-101. [DOI] [PubMed] [Google Scholar]
- 20.Zhang W, Hawse J, Huang Q, Sheets NL, Hosack A. Lempicki RA, Horwitz J, Kantorow M. Decreased expression of ribosomal proteins in human age-related cataract. Invest Ophthalmol Vis Sci. 2002;43(1):198–204. [PMC free article] [PubMed] [Google Scholar]
- 21.Kondoh N, Noda M, Fisher RJ, Schweinfest CW, Papas TS, Kondoh A, Samuel KP, Oikawa T. The S29 ribosomal protein increases tumor suppressor activity of K rev-1 gene on v-K ras-transformed NIH3T3 cells. Biochim Biophys Acta. 1996;1313(1):41–46. doi: 10.1016/0167-4889(96)00052-3. [DOI] [PubMed] [Google Scholar]
- 22.Khanna N, Reddy VG, Tuteja N, Sinqh N. Differential gene expression in apoptosis: identification of ribosomal protein S29 as an apoptotic inducer. Biochem Biophys Res Commun. 2000;277(2):476–486. doi: 10.1006/bbrc.2000.3688. [DOI] [PubMed] [Google Scholar]
- 23.Tang SJ, Wang XK, Chen JH, Wang XY, Liang WZ, Liang XQ. Effect of ribosomal protein s29 gene expression product on the proliferation of skin fibroblasts in non-scar healing. Chin J Clin Rehabil. 2004;8(23):4762–4764. [Google Scholar]
- 24.Cavdar Koc E, Ranasinghe A, Burkhart W, Blackburn K, Koc H, Moseley A, Spremulli LL. A new face on apootosis: death-associated protein 3 and PDCD9 are mitochondrial ribosomal proteins. FEBS Letts. 2003;492(1–2):166–170. doi: 10.1016/s0014-5793(01)02250-5. [DOI] [PubMed] [Google Scholar]


