Rapamycin modulates the ability of the vascular endothelium to mediate inflammation by inhibiting mTORC2 and reducing TNF-induced VCAM-1 expression.
Abstract
Recruitment of circulating leukocytes into inflamed tissues depends on adhesion molecules expressed by endothelial cells (ECs). Here we report that rapamycin pretreatment reduced the ability of TNF-treated ECs to capture T cells under conditions of venular flow. This functional change was caused by inhibition of TNF-induced expression of vascular cell adhesion molecule-1 (VCAM-1) and could be mimicked by knockdown of mammalian target of rapamycin (mTOR) or rictor, but not raptor, implicating mTORC2 as the target of rapamycin for this effect. Mechanistically, mTORC2 acts through Akt to repress Raf1-MEK1/2-ERK1/2 signaling, and inhibition of mTORC2 consequently results in hyperactivation of ERK1/2. Increased ERK1/2 activity antagonizes VCAM-1 expression by repressing TNF induction of the transcription factor IRF-1. Preventing activation of ERK1/2 reduced the ability of rapamycin to inhibit TNF-induced VCAM-1 expression. In vivo, rapamycin inhibited mTORC2 activity and potentiated activation of ERK1/2. These changes correlated with reduced endothelial expression of TNF-induced VCAM-1, which was restored via pharmacological inhibition of ERK1/2. Functionally, rapamycin reduced infiltration of leukocytes into renal glomeruli, an effect which was partially reversed by inhibition of ERK1/2. These data demonstrate a novel mechanism by which rapamycin modulates the ability of vascular endothelium to mediate inflammation and identifies endothelial mTORC2 as a potential therapeutic target.
In the presence of the inflammatory cytokine TNF, endothelial cells (ECs) express adhesion molecules that facilitate recruitment of leukocytes to damaged tissues (Pober and Sessa, 2007). Vascular cell adhesion molecule-1 (VCAM-1) is a TNF-inducible adhesion molecule that facilitates capture of leukocytes expressing VCAM-1 counter-receptors, α4β1 and α4β7 integrins (Cook-Mills et al., 2011). VCAM-1 is expressed in inflammatory disorders, and its blockade reduces disease in models of multiple sclerosis, inflammatory bowel disease, and asthma (Cook-Mills et al., 2011). Thus, the ability to modulate VCAM-1 expression may be of therapeutic interest.
TNF activates signaling cascades including the MAP kinase (p38, JNK1/2, and ERK1/2) and NF-κB pathways (Madge and Pober, 2001). TNF signaling drives VCAM-1 expression by activating AP-1, NF-κB, and IRF-1 transcription factors (Ahmad et al., 1995; Lechleitner et al., 1998; Tsoyi et al., 2010). After ligation of TNF receptor 1, Raf1 is recruited to and activated at the cell membrane in a manner dependent on the adaptor protein TRADD (Xu et al., 1998). Activated Raf1 phosphorylates and activates the MEK1/2 kinases, which in turn dually phosphorylate and activate ERK1/2 (Roberts and Der, 2007). The role of ERK1/2 in relation to VCAM-1 expression is unclear.
Rapamycin is a macrolide antibiotic that inhibits mammalian target of rapamycin (mTOR), a protein kinase which nucleates two distinct signaling complexes, known as mTORC1 and mTORC2 (Laplante and Sabatini, 2012). mTORC1 contains the binding partner raptor and is inhibited acutely (in minutes) by rapamycin, whereas mTORC2, which associates with rictor, is only disrupted after more prolonged (24 h) treatment (Sarbassov et al., 2006). Active mTORC2 phosphorylates the hydrophobic motif of Akt (Ser473), resulting in its full activation; defective phosphorylation at this site impairs the ability of Akt to phosphorylate a subset of its targets (Jacinto et al., 2006).
In a humanized mouse model of transplantation, we observed that human arterial allografts pretreated with rapamycin contained fewer alloreactive T cells (Wang et al., 2013) and wondered whether rapamycin interfered with EC recruitment of leukocytes. Here we show that prolonged rapamycin pretreatment reduced the capacity of TNF-treated ECs to capture leukocytes under conditions of venular flow. This effect is causally linked to inhibition of mTORC2, resulting in diminished TNF-induced VCAM-1 expression. Mechanistically, inhibition of mTORC2 leads to hyperactivation of ERK1/2, which in turn reduces TNF-induced VCAM-1 expression by repressing induction of the transcription factor IRF-1. In vivo, rapamycin inhibited mTORC2 activity, potentiated activation of ERK1/2, reduced endothelial expression of TNF-induced VCAM-1, and decreased infiltration of leukocytes into renal glomeruli. Both in vitro and in vivo inhibition of ERK1/2 reversed the inhibitory actions of rapamycin.
RESULTS AND DISCUSSION
Rapamycin reduces T cell capture and VCAM-1 expression by TNF-activated ECs via inhibition of mTORC2
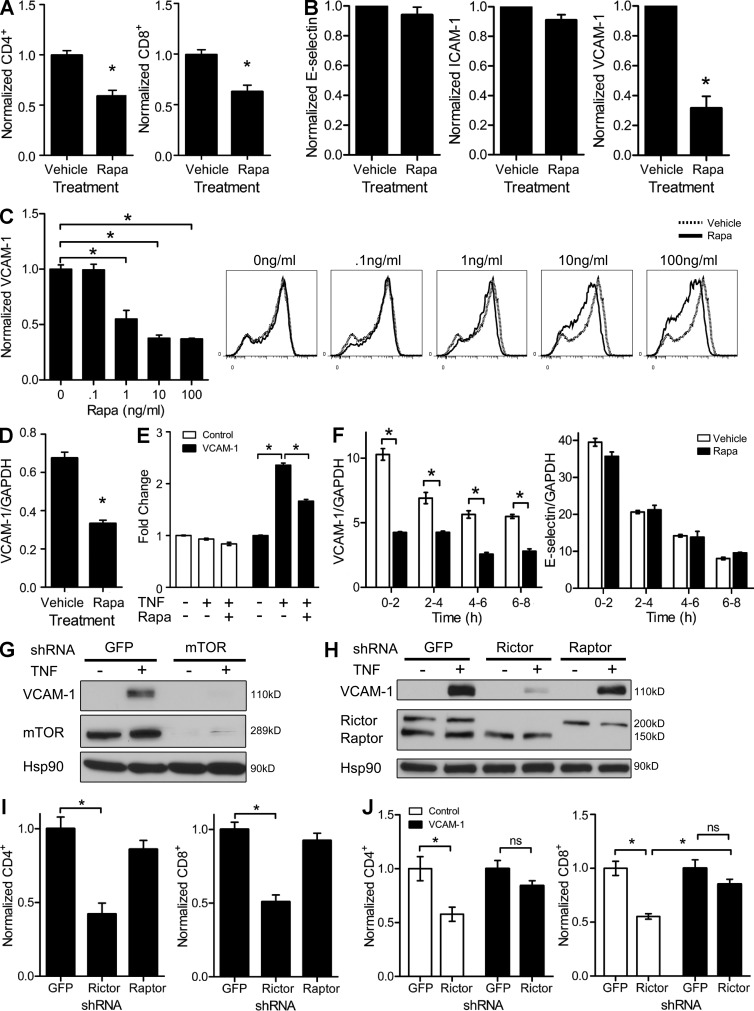
We tested the ability of TNF-activated control and rapamycin-treated ECs (rapa-ECs) to capture T cells and found that fewer human memory CD8 and CD4 T cells adhered to rapa-ECs under conditions of venular shear stress (Fig. 1 A). Because T cell recruitment is dependent on cytokine-inducible adhesion molecules, we investigated whether rapamycin affected expression of such molecules. We found no differences in induction of E-selectin or ICAM-1 (Fig. 1 B). However, rapamycin significantly reduced TNF-induced VCAM-1 expression in a dose-dependent manner (Fig. 1, B and C). TNF-activated rapa-ECs expressed lower levels of VCAM-1 mRNA (Fig. 1 D), and rapamycin inhibited luciferase expression driven by a 2-kb region of the VCAM-1 promoter containing NF-κB–, AP-1–, and IRF-1–binding sites (Fig. 1 E). Consistent with the response of the promoter-reporter gene, rapamycin decreased transcription of the endogenous vcam-1 gene in response to TNF; this effect appeared specific in that transcription of the gene encoding E-selectin (Sele) was not inhibited (Fig. 1 F).
Figure 1.
Rapamycin reduces VCAM-1 expression by inhibiting mTORC2. (A) TNF-activated control- or rapa-ECs were subjected to adhesion assays using human memory T cells. (B) TNF-activated control- and rapa-ECs were analyzed by FACS. (C) ECs pretreated with rapamycin were treated with TNF and assayed by FACS. Data are normalized to median fluorescence intensity (MFI) of staining in control-ECs. (D) VCAM-1 mRNA expression in control- or rapa-ECs stimulated with TNF. (E) Control- or rapa-ECs transduced with the indicated luciferase reporters were stimulated with TNF and assayed for luciferase expression. Values are normalized to expression in control-ECs (no TNF, no rapamycin). (F) Quantification of nascent VCAM-1 and E-selectin mRNA expression in control- and rapa-ECs stimulated with TNF. (G) TNF-activated GFP- or mTOR-knockdown ECs were assayed by Western blotting. (H) TNF-activated GFP-, rictor-, or raptor-knockdown ECs were assayed by Western blotting. (I) TNF-activated GFP-, rictor-, or raptor-knockdown ECs were subjected to adhesion assay. (J) TNF-activated control or VCAM-1–overexpressing ECs were transduced with GFP or rictor shRNA and subjected to adhesion assay. For G and H, similar results were seen in two independent experiments. Mean ± SEM are shown for data pooled from two (A [n = 6], C [n = 4], E [n = 6], F [n = 5], I [n = 6], and J [n = 6]) or three (B [n = 3] and D [n = 4]) independent experiments (n = total replicates). For C, E, and J, significance was determined by ANOVA with Tukey’s post-hoc test; all other data were analyzed with the Student’s t test. *, P < 0.05.
To confirm the specificity of rapamycin, we silenced mTOR using shRNA. Similar to rapamycin, mTOR knockdown decreased levels of VCAM-1 after TNF stimulation (Fig. 1 G). In addition, knockdown of rictor, but not raptor, reduced TNF-induced VCAM-1 (Fig. 1 H), implicating mTORC2 as the target of rapamycin for this effect. Functionally, knockdown of rictor reduced the ability of ECs to capture T cells (Fig. 1 I), an effect which was significantly reversed by overexpression of VCAM-1 (Fig. 1 J).
Inhibition of mTORC2 reduces VCAM-1 expression by potentiating activation of ERK1/2
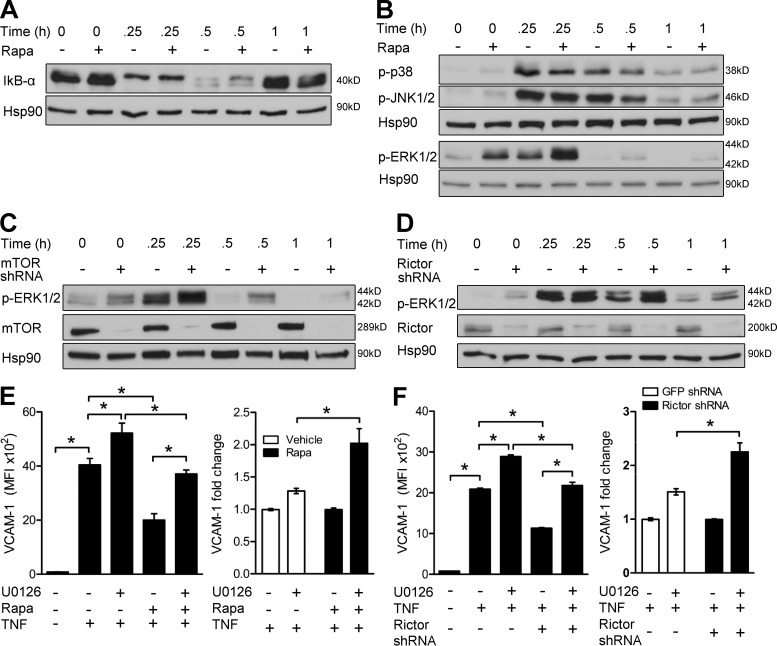
We next investigated whether TNF signaling pathways were altered by rapamycin. There were no differences in the processing of IκB-α, the degradation of which is linked to NF-κB activation (Fig. 2 A), or phosphorylation of p38 MAPK and JNK1/2, upstream activators of AP-1 (Fig. 2 B), in TNF-activated control or rapa-ECs. However, rapa-ECs displayed heightened basal and TNF-induced activation of ERK1/2 (Fig. 2 B). A similar effect was seen in mTOR (Fig. 2 C)- and rictor-knockdown ECs (Fig. 2 D), suggesting that rapamycin inhibition of mTORC2 causes hyperactivation of ERK1/2. A rapid decline in TNF-induced ERK phosphorylation at 30 and 60 min likely represents ERK-mediated negative feedback (Fritsche-Guenther et al., 2011).
Figure 2.
Rapamycin inhibition of mTORC2 reduces VCAM-1 expression by hyperactivating ERK1/2. (A and B) Western blot analysis of control- or rapa-ECs stimulated with TNF for the indicated times. (C and D) Western blot analysis of ECs transduced with the indicated shRNA and treated with TNF for the indicated times. (E) ECs pretreated with rapamycin and/or U0126 were stimulated with TNF and analyzed by FACS. Left panel shows MFI of VCAM-1 staining. Right panel shows induction of VCAM-1 by U0126 relative to vehicle- or rapa-ECs not exposed to U0126. (F) ECs transduced with the indicated shRNA were pretreated with U0126, stimulated with TNF, and then analyzed by FACS. Left panel shows MFI of VCAM-1 staining in a representative experiment (n = 3). Right panel shows pooled data comparing fold induction of VCAM-1 by U0126 relative to control- or rictor-knockdown ECs not exposed to U0126. For A–D, similar results were seen in two independent experiments. Means ± SEM are shown for data pooled from two (E [n = 9] and F, right [n = 6]) independent experiments (n = total replicates). For E and F, significance was determined by ANOVA with Tukey’s post-hoc test. *, P < 0.05.
To interrogate the role of ERK1/2, we inhibited ERK1/2 activation using an MEK1/2 inhibitor (U0126) and found that U0126 partially restored TNF-induced VCAM-1 expression in both rapa-ECs (Fig. 2 E) and rictor-knockdown ECs (Fig. 2 F). Although inhibition of ERK1/2 increased VCAM-1 expression in all groups, the relative increase was much greater in rapa-ECs (Fig. 2 E) and rictor-knockdown ECs (Fig. 2 F). The variation in the absolute expression levels of VCAM-1 in these experiments is likely caused by the genetic diversity among cultures of ECs. However, the relative reductions in TNF-induced VCAM-1 by rapamycin and rictor knockdown are similar in magnitude. These data suggest that mTORC2 represses activation of ERK1/2 so that inhibition of mTORC2 by rapamycin or rictor knockdown consequently leads to increased ERK1/2 activity, which acts to inhibit VCAM-1 expression.
The mTORC2 target Akt antagonizes Raf1-MEK1/2-ERK1/2 signaling
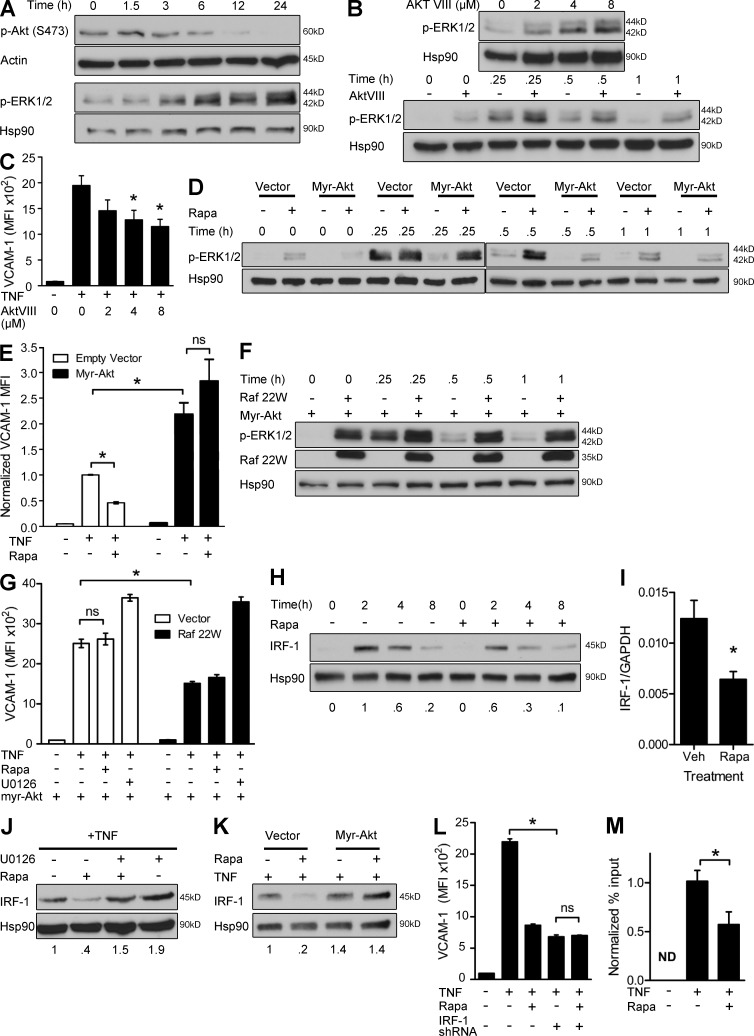
Rapamycin reduced mTORC2 phosphorylation of Akt, and the kinetics of this reduction correlated with increasing levels of phospho-ERK1/2 (Fig. 3 A). To determine whether mTORC2-dependent Akt activity represses ERK1/2 and thereby promotes TNF induction of VCAM-1, we inhibited Akt with a pharmacological inhibitor (AktVIII). AktVIII increased basal and TNF-induced phosphorylation of ERK1/2 (Fig. 3 B) and decreased TNF-induced VCAM-1 expression (Fig. 3 C). Conversely, expression of constitutively active myristoylated Akt diminished activation of ERK1/2 (Fig. 3 D), potentiated induction of VCAM-1, and rendered VCAM-1 expression resistant to the effects of rapamycin (Fig. 3 E). We conclude that rapamycin inhibits mTORC2-dependent Akt activity, leading to hyperactivation of ERK1/2 and diminished TNF-induced VCAM-1.
Figure 3.
mTORC2-dependent Akt signaling inhibits Raf1-MEK1/2-ERK1/2, leading to defective induction of IRF-1. (A) Western blot analysis of ECs treated with rapamycin for the indicated times. (B, top) Western blot analysis of ECs treated with the indicated doses of AktVIII for 24 h. (bottom) Western blot analysis of ECs treated with 4 µM AktVIII for 24 h and then stimulated with TNF for the indicated times. (C) ECs were pretreated with the indicated doses of AktVIII and then stimulated with TNF overnight. VCAM-1 expression was assessed by FACS. (D) ECs transduced with the indicated constructs were pretreated with rapamycin, stimulated with TNF for the indicated times, and assayed by Western blotting. (E) ECs transduced with the indicated constructs were pretreated with rapamycin and stimulated with TNF and then assayed by FACS. Data are normalized to MFI of VCAM-1 staining in empty vector–ECs treated with TNF. (F) ECs transduced with the indicated constructs were stimulated with TNF for the indicated times and assayed by Western blotting. (G) ECs expressing myristoylated Akt were transduced with control or Raf 22W vector. Transduced ECs were pretreated with rapamycin or U0126 and stimulated overnight with TNF. VCAM-1 expression was assessed by FACS. (H) Western blot analysis of control- or rapa-ECs stimulated with TNF for the indicated times. (I) mRNA expression in control- and rapa-ECs stimulated with TNF for 0.25 h. Veh, vehicle. (J) Western blot analysis of ECs pretreated with rapamycin and/or U0126 and then stimulated with TNF for 2 h. (K) ECs transduced with the indicated constructs were pretreated with rapamycin, stimulated with TNF for 2 h, and assayed by Western blotting. (L) ECs were transduced with control or IRF-1 shRNA vectors, as indicated. Transduced ECs were then pretreated with rapamycin and stimulated overnight with TNF, as indicated. VCAM-1 expression was assessed by FACS after TNF stimulation. (M) ChIP analysis of control and rapa-ECs stimulated with TNF for 2 h. DNA quantification is expressed as percent input and normalized to TNF treatment sample. ND, not detected. For A, B, D, F, H, J, and K, similar results were seen in two independent experiments. Mean ± SEM are shown for data pooled from two (C [n = 6], E [n = 6], G [n = 6], I [n = 3], L [n = 6], and M [n = 4]) independent experiments (n = total replicates). For E, G, and L, significance was determined by ANOVA with Tukey’s post-hoc test; all other data were analyzed with the Student’s t test. *, P < 0.05.
ERK1/2 is phosphorylated by MEK1/2, which is phosphorylated by Raf1. Akt is known to antagonize Raf1, and such a link has been shown in ECs (Ren et al., 2010). To determine the relation between Akt and Raf1, we expressed a constitutively active Raf1 mutant (Raf 22W) in ECs that also express myristoylated Akt. We found that even in the setting of constitutive Akt activity, coexpressed Raf 22W activated ERK1/2 (Fig. 3 F) and decreased TNF-induced VCAM-1 (Fig. 3 G). These data place Raf1 downstream of Akt, implying that Akt inhibits ERK1/2 activation at the level of Raf1 or higher.
Rapamycin reduces VCAM-1 expression by inhibiting TNF induction of IRF-1
The TNF-induced transcription factor IRF-1 promotes VCAM-1 transcription (Lechleitner et al., 1998). Rapamycin reduced IRF-1 protein and mRNA after TNF stimulation (Fig. 3, H and I). Defective induction of IRF-1 reduced VCAM-1 expression, which could be restored in rapa-ECs by inhibition of ERK1/2 (Fig. 3 J) or transduction with myristoylated Akt (Fig. 3 K). Furthermore, shRNA knockdown of IRF-1 reduced TNF-induced VCAM-1 expression and abolished the ability of rapamycin to further decrease VCAM-1 expression (Fig. 3 L). Lastly, TNF-induced binding of IRF-1 to the VCAM-1 promoter, as assessed by chromatin immunoprecipitation (ChIP), was reduced by rapamycin (Fig. 3 M). We conclude that hyperactivation of ERK1/2 by rapamycin reduces TNF-induced VCAM-1 by impairing induction of IRF-1.
Rapamycin increases ERK1/2 activation and reduces TNF-induced VCAM-1 expression in vivo
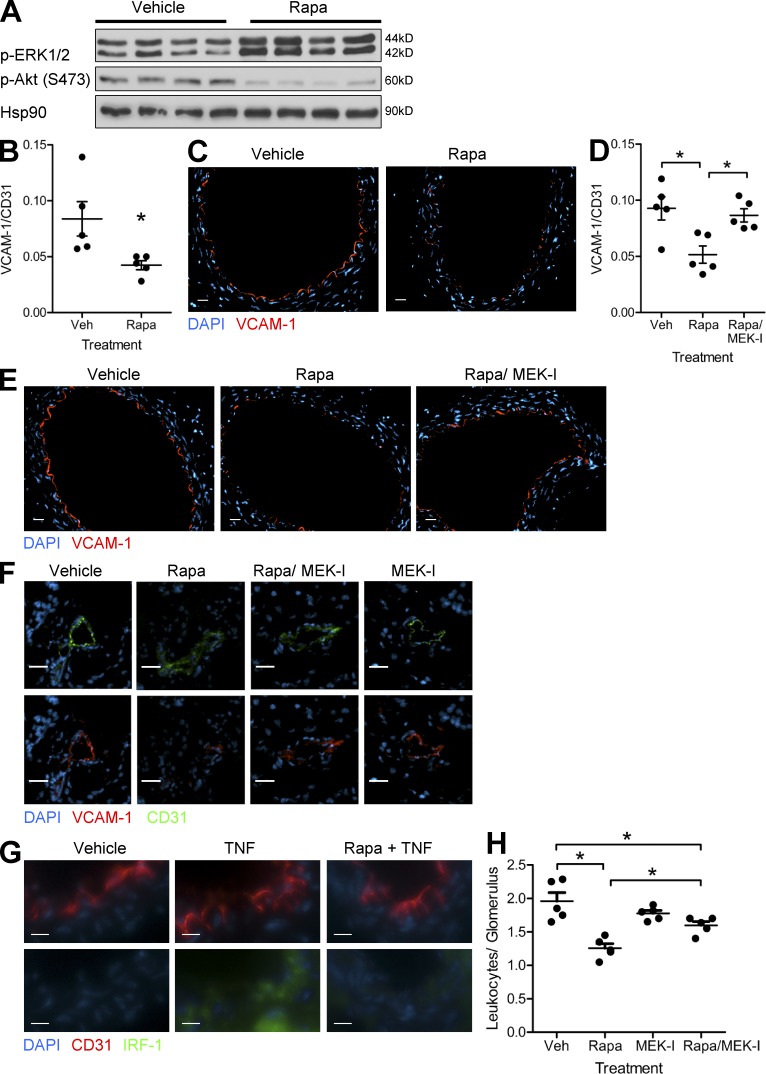
To determine the relevance of our findings, we treated mice with rapamycin before injection with TNF. We found that rapamycin inhibited mTORC2-mediated phosphorylation of Akt in murine aortas (Fig. 4 A), which coincided with increased activation of ERK1/2 (Fig. 4 A) and reduced VCAM-1 mRNA transcripts (Fig. 4 B). By immunofluorescence, TNF-induced VCAM-1 expression was only observed on ECs in the aorta and was diminished in mice treated with rapamycin (Fig. 4 C). Pharmacological inhibition of ERK1/2 with an MEK1/2 inhibitor (PD0325901) restored TNF-induced VCAM-1 mRNA (Fig. 4 D) and protein (Fig. 4 E) expression in mice treated with rapamycin.
Figure 4.
Rapamycin reduces VCAM-1 expression in vivo. (A) Mice were pretreated with vehicle or rapamycin and then injected with TNF. Aortas were harvested after 16 h and assayed by Western blotting. Lanes represent individual mice. (B) Expression of VCAM-1 mRNA (normalized to CD31) in aorta from mice treated as in A (n = 5 per group). (C) Mice were treated as in A. Harvested aortas were analyzed for VCAM-1 expression via immunofluorescence. (D) Expression of VCAM-1 mRNA (normalized to CD31) in aortas harvested from mice pretreated with vehicle, rapamycin, or rapamycin + MEK inhibitor (MEK-I; PD0325901) and then injected with TNF (n = 5 per group). (E) Mice were treated as in D. Harvested aortas were analyzed for VCAM-1 expression via immunofluorescence. (F) VCAM-1 expression in renal microvessels from mice pretreated with vehicle, rapamycin, rapamycin + MEK inhibitor, or MEK inhibitor and then injected with TNF. (G) IRF-1 expression in renal microvessels from mice pretreated with vehicle and rapamycin and then injected with TNF. (C and E–G) Representative images are shown. Bars: (C, E, and F) 25 µm; (G) 10 µm. (H) Leukocyte infiltration into renal glomeruli in mice treated as in F (n = 5 per group). Veh, vehicle. For B, D, and H, mean ± SEM is shown. For D and H, significance was determined by ANOVA with Tukey’s post-hoc test; all other data were analyzed with the Student’s t test. *, P < 0.05.
To assess the functional effects of these changes, we analyzed TNF-induced infiltration of leukocytes. Although C.B-17 SCID/bg mice lack T cells, they have circulating monocytes that, like T cells, bind to VCAM-1. We found that TNF induces VCAM-1 expression in cardiac, hepatic, and renal microvessels and that VCAM-1 induction is associated with sparse but measurable leukocyte infiltration into these sites (not depicted). We focused on leukocyte recruitment to renal glomeruli as infiltration at this site is most easily quantified. Rapamycin diminished TNF-induced VCAM-1 expression in renal microvessels and pharmacological inhibition of ERK1/2 restored this expression (Fig. 4 F). Rapamycin also reduced TNF-induced IRF-1 expression in renal endothelium (Fig. 4 G). We found that rapamycin reduced the number of CD45+ leukocytes infiltrating into glomeruli after TNF treatment and that inhibition of ERK1/2 partially reversed this effect (Fig. 4 H). Interpretation of this experiment is complicated because we cannot separate the effects of rapamycin and PD0325901 on ECs from their effects on leukocytes. However, these data are consistent with our in vitro observations on the effects of these agents on VCAM-1 expression and T cell capture.
The endothelium is a key mediator of inflammation and immunity (Pober and Sessa, 2007). Inappropriate activation of these functions results in excessive inflammation and is thought to play a pathological role in atherosclerosis (O’Brien et al., 1993), rheumatoid arthritis (Koch et al., 1991), and cardiac allograft vasculopathy (Tellides and Pober, 2007). The majority of antiinflammatory therapies are targeted at the immune system itself. Rapamycin was initially introduced as an agent for controlling transplant rejection as the result of its ability to limit T cell proliferation. Because of the essential roles played by vascular endothelium in inflammation (Al-Lamki et al., 2008), targeting the endothelium may provide an alternative approach to limiting pathological processes, and many agents that act on the immune system may also have effects on the endothelium. This insight was supported by our recent observation that pretreatment of human arterial allograft ECs with rapamycin protected against subsequent T cell–mediated rejection (Wang et al., 2013). This finding prompted us to investigate the effects of rapamycin on EC recruitment of leukocytes. Indeed, we find that rapamycin inhibits the ability of TNF-activated ECs to capture leukocytes under conditions of venular flow in vitro and reduces infiltration of leukocytes into renal glomeruli in vivo. These effects correlated with inhibition of mTORC2 and reduced induction of VCAM-1 on rapa-ECs. Although the inhibitory effects of rapamycin on leukocyte recruitment are not complete, they are significant and likely contribute to the protective effects of rapamycin on allografts. Moreover, the experiments reported here demonstrate a novel signaling cascade through which rapamycin can modulate the ability of vascular endothelium to mediate inflammation and identify endothelial mTORC2 as a potential therapeutic target.
MATERIALS AND METHODS
Isolation and culture of human cells.
All human cells were obtained under protocols approved by the Institutional Review Board of Yale University. PBMCs were isolated by density centrifugation of leukopheresis products obtained from anonymized healthy adult volunteers. CD4+ or CD8+ T cells were isolated from PBMCs using Dynabeads (Invitrogen) magnetic beads, according to manufacturer’s instructions. Activated T cells and monocytes were depleted by incubating the isolated cells with mouse anti-CD14 (BioLegend) and anti–HLA-DR (LB3.1; gift of J. Strominger, Harvard University, Cambridge, MA) antibodies, followed by incubation with magnetic pan-mouse IgG beads (Invitrogen). Memory T cells were isolated by further incubating T cells with mouse anti-CD45RA (eBioscience) and pan-mouse IgG beads. Isolates were routinely >99% CD4+ HLA-DR−CD45RA− by flow cytometry.
Human umbilical vein ECs were isolated from umbilical cords by collagenase digestion. ECs were serially cultured on 0.1% gelatin–coated tissue culture plates in M199 (Invitrogen) supplemented with 20% FBS, 2 mM l-glutamine, 100 U/ml penicillin, 100 µg/ml streptomycin, 0.1% EC growth supplement (Collaborative Biomedical Research), and 100 µg/ml porcine heparin (Sigma-Aldrich). EC cultures were used at passage levels 2–5, at which time the cultured cells are uniformly positive for the EC marker CD31 and are devoid of CD45+ contaminating leukocytes.
In vitro TNF and pharmacological inhibitor treatments.
Confluent ECs were treated with vehicle (DMSO), rapamycin (Sigma-Aldrich), U0126 (Cell Signaling Technology), or AktVIII (EMD Millipore) as indicated for 24 h, at which time fresh media containing the same inhibitor and 10 ng/ml TNF (R&D Systems) was added. For FACs experiments, ECs were analyzed after 6 (E-selectin) or 16 h (VCAM-1 and ICAM-1) of TNF treatment. For quantitative real-time RT-PCR experiments, ECs were analyzed after 16 h of TNF treatment. Unless otherwise noted, inhibitors were used at the following concentrations: 100 ng/ml rapamycin, 10 µM U0126, and 8 µM AktVIII. In experiments examining phosphorylation of MAP kinase proteins in response to TNF, inhibitor and cytokine treatments were performed in M199 media containing 1% FBS, 2 mM l-glutamine, 100 U/ml penicillin, 100 µg/ml streptomycin, and porcine heparin. ECs were harvested after TNF treatment at the indicated time points.
FACS analysis.
ECs were washed with HBSS, suspended with trypsin, washed in PBS, and stained (on ice) in PBS supplemented with 1% BSA with PE-conjugated anti–human ICAM1, PE-conjugated anti–human E-selectin, or FITC-conjugated anti–human VCAM-1 (all from BD). After 30 min, cells were washed twice in PBS and analyzed on an LSRII flow cytometer (BD). FACS data were analyzed and interpreted using FlowJo software (Tree Star).
Immunoblotting.
Cultured ECs were washed with ice-cold PBS and lysed directly by addition of NP-40 lysis buffer (50 mM Tris-HCl, 10% glycerol, 125 mM NaCl, 1% NP-40, 5.3 mM NaF, 1 mM orthovanadate, and 1 mg/ml protease inhibitor mixture [Roche]). Cell lysates were collected, subjected to freeze-thaw, sonicated, and then cleared by centrifugation at 12,000 g for 10 min. For in vivo experiments, thoracic aortas were harvested from mice and flash frozen in liquid nitrogen. To extract protein, aortas were thawed on ice, suspended in NP-40 lysis buffer, sonicated, and then cleared by centrifugation at 12,000 g for 10 min. Protein concentrations of all samples were measured using DC Protein Assay (Bio-Rad Laboratories), and equal amounts of protein from each sample were boiled at 95°C in SDS sample buffer for 10 min. Proteins were separated by electrophoresis on a 10% SDS-PAGE gel and transferred to PVDF membrane (EMD Millipore) at 100 V for 1 h at 4°C. After blocking in TBST containing 5% BSA, membranes were incubated overnight at 4°C with antibodies against mTOR, rictor, raptor, phospho-ERK1/2, phospho-p38, phospho-JNK, phospho-S6K, phospho-Akt (S473), Hsp90, IκB-α, Raf1, IRF-1 (all from Cell Signaling Technology), actin (Sigma-Aldrich), or VCAM-1 (R&D Systems). Bound antibodies were detected with HRP-conjugated goat anti–mouse or goat anti–rabbit secondary antibodies (Jackson ImmunoResearch Laboratories, Inc.) and SuperSignal West Pico chemiluminescent substrate (Thermo Fisher Scientific).
Retrovirus generation and transduction.
mTOR, raptor, rictor, IRF-1, or GFP (control) was silenced in ECs via lentiviral shRNA. To generate virus, 293T cells were transfected, using Lipofectamine 2000 (Invitrogen), with plasmids encoding shRNA targeting mTOR, raptor, rictor, IRF-1, or GFP together with CMV-dR8.2 packaging and CMV-VSVG envelope plasmids. Supernatants containing virus were collected 48 and 72 h after transfection and used to infect ECs in the presence of 8 µg/ml polybrene. ECs underwent two rounds of transduction, and transduced ECs were selected via puromycin resistance.
To generate retrovirus encoding myristoylated Akt, Raf 22W, VCAM-1, or empty vector (pBABE puro or pLZRS), Phoenix cells (gift of G. Nolan, Stanford University, Stanford, CA) were transfected, using Lipofectamine 2000, with plasmids encoding myristoylated Akt, Raf 22W, VCAM-1 cDNA, or empty vector. Viral supernatants were collected and used to transduce ECs as described above. mTOR shRNA (Addgene plasmid 1856), raptor shRNA (Addgene plasmid 1858), rictor shRNA (Addgene plasmid 1853), and GFP shRNA (Addgene plasmid 30323) plasmids were deposited by D. Sabatini (Massachusetts Institute of Technology [MIT], Cambridge, MA). CMV-dR8.2 and CMV-VSVG (Addgene plasmids 8455 and 8454) plasmids were deposited by R. Weinberg (MIT). Myristoylated Akt (Addgene plasmid 15294) plasmid was deposited by W. Hahn (Dana-Farber Cancer Institute, Boston, MA). Raf 22W (Addgene plasmid 12593) plasmid was deposited by C. Der (The University of North Carolina at Chapel Hill, Chapel Hill, NC). VCAM-1 overexpression vector was previously described (Manes et al., 2006).
T cell adhesion assay.
ECs were treated with vehicle or rapamycin for 24 h, at which time they were collected by trypsinization and plated at confluency into 35-mm dishes. In some experiments GFP-, rictor-, or raptor-knockdown ECs were plated, whereas in others VCAM-1–overexpressing ECs transduced with GFP or rictor shRNA were plated. After ECs attached to the dish, they were activated with TNF for 16 h and tested in adhesion assays. To assess adhesion under conditions of venular shear stress, a parallel plate flow chamber apparatus (Glycotech) was mounted on the EC monolayer placed on a 37°C heating surface. Human T cells (106 cells/500 µl), prestained with anti-CD45 (BioLegend) followed by Alexa Fluor 488–conjugated goat anti–mouse IgG (Invitrogen), suspended in RPMI/10% FBS, were loaded onto the EC monolayer at 0.75 dyn/cm2 for 2 min, followed by washing with medium only at 1 dyn/cm2 for 5 min. Samples were then fixed with 3.7% formaldehyde in PBS, mounted on slides using mounting medium (Prolong Gold; Invitrogen), and examined by microscopy with an Axiovert 200M microscope (Carl Zeiss). An FITC filter was used to detect Alexa Fluor 488–stained cells. CD45-positive T cells in 10 fields (1,000 × 1,000 pixels with a 10× objective) covering the length of the sample were counted. Triplicate dishes were counted for each experimental group.
Promoter-reporter gene assay.
VCAM-1 transcription was examined using a lentiviral transcriptional reporter system (System Biosciences). To make this reporter, a 2-kb region of the vcam-1 promoter containing AP-1, NF-κB, and IRF-1 sites was PCR amplified from genomic DNA, with ClaI and SpeI restriction sites introduced at either end of the promoter region. This PCR product was directionally cloned into (promoterless) pGreenFire vector upstream of the luciferase-encoding region. All constructs were confirmed by sequencing. To generate retrovirus, phoenix cells were transfected, using Lipofectamine 2000, with the constructed plasmid or an empty vector. Viral supernatants were collected and used to transduce ECs as described in the section above. Transduced ECs were treated with 100 ng/ml rapamycin for 24 h, followed by 10 ng/ml TNF for 6 h, at which time cells were lysed. Luciferase activity in lysates was assayed using the Firefly Luciferase Assay System (Promega) and quantified with a Lumat LB 9501 luminometer (Berthold).
Analysis of endogenous gene transcription.
Analysis of transcription from the vcam-1 and sele genes, encoding VCAM-1 and E-selectin, respectively, was performed using the Click-iT Nascent RNA capture kit (Invitrogen), according to the manufacturer’s instructions. In brief, confluent cultured ECs were treated with 0 or 100 ng/ml rapamycin for 24 h, at which time they were stimulated with 10 ng/ml TNF for 8 h. During this stimulation period, separate EC plates were pulsed with 0.2 mM 5-ethynyl uridine (EU) at 2-h intervals (0–2, 2–4, 4–6, and 6–8 h); all ECs were collected and flash frozen after stimulation. Total RNA was isolated from collected cells using the RNeasy Mini kit (QIAGEN), according to the manufacturer’s instructions. Click reaction was performed on this isolated RNA to biotinylate any incorporated EU residues. Biotinylated RNA was then isolated using Dynabeads MyOne Streptavidin T1 magnetic beads (Invitrogen). Isolated RNA was reverse transcribed using the High-Capacity cDNA Reverse Transcription kit (Applied Biosystems). The resulting cDNA was analyzed via quantitative real-time RT-PCR analysis, as described below.
ChIP.
ChIP was performed as previously described (Sun et al., 2012) using the ChIP-IT Express enzymatic kit (Active Motif). In brief, ECs were treated with 0 or 100 ng/ml rapamycin for 24 h and then stimulated with 10 ng/ml TNF for 2 h. After stimulation, ECs were fixed with 1% formaldehyde for 10 min. Fixation was stopped using glycine stop-fix solution, and cells were harvested by scraping. Collected cells were lysed using lysis buffer and passing cells through a 25-gauge needle. Cell nuclei were isolated by centrifugation at 5,000 rpm for 10 min, resuspended in 350 µl digestion buffer, and digested using 17 µl of enzymatic shearing cocktail for 10 min. Digestion was stopped by adding 7 µl of 0.5M EDTA to the reaction. An aliquot of the digested chromatin was measured with a NanoDrop Spectrophotometer and run on a 1.5% agarose gel to ensure proper digestion to 200–1,000-bp fragments. For immunoprecipitation, chromatin corresponding to 10 µg DNA was incubated with protein G magnetic beads and 3 µg rabbit anti–human IRF-1 (Santa Cruz Biotechnology, Inc.) or control rabbit IgG (Cell Signaling Technology) overnight at 4°C. After extensive washing of beads, bound chromatin was eluted and DNA–protein cross-links were reversed. DNA was subsequently purified using the Chromatin IP DNA Purification kit (Active Motif) and analyzed via quantitative real-time RT-PCR as described below. Quantitative real-time RT-PCR was performed using iQ SYBR Green Supermix (Bio-Rad Laboratories) with the following primers designed to amplify the IRF-1–binding site within the vcam-1 promoter: R, 5′-GTGAGGCCCGATGCAGATA-3′; and F, 5′-GAACTTGGCTGGGTGTCTGTTA-3′. Cycling conditions were as follows: 95°C for 10 min followed by 40 cycles of 95°C for 15 s and 61°C for 50 s. Data were analyzed and expressed as percent input and subsequently normalized to the TNF-treated sample.
In vivo mouse experiments.
All experimental animal protocols were approved by the Yale University Institutional Animal Care and Use Committee. Rapamycin (sirolimus; EMD Millipore) and MEK inhibitor PD0325901 (Selleckchem) were diluted in ethanol and mixed with an equal volume (30 µl) of Cremophor EL (Sigma-Aldrich). These drug mixtures were dissolved in Dulbecco’s PBS at a final volume of 0.2 ml/mouse and administered via intraperitoneal injection. Female C.B-17 SCID/beige mice (Taconic) mice were injected with 3 mg/kg sirolimus, 25 mg/kg PD0325901, or 3 mg/kg sirolimus + 25 mg/kg PD0325901 every 24 h for 3 d. Control mice were injected with a mixture containing ethanol and Cremophor EL in PBS. After the final rapamycin treatment, mice were injected with 30 µg/kg recombinant human TNF (R&D Systems) diluted in 0.2 ml PBS via the tail vein. After 16 h, mice were anesthetized and perfused with saline, and portions of the aorta, kidney, liver, and heart were harvested. Tissue samples were then either flash frozen in liquid nitrogen or frozen in optimal cutting temperature (OCT) compound (Sakura). Serial 5- or 30-µm transverse sections were cut for immunofluorescence or quantitative real-time RT-PCR analysis, respectively.
Quantitative real-time RT-PCR analysis.
To isolate RNA from ECs, cultured cells were washed with HBSS, collected by trypsinization, and processed using the RNeasy Mini kit (QIAGEN). To isolate RNA from mouse tissue, serial 30-µm sections of frozen tissue embedded in OCT were cut, collected, and immersed in RLT lysis buffer (QIAGEN) and then processed using the RNeasy Mini kit. Isolated RNA was converted to cDNA using the High-Capacity cDNA Reverse Transcription kit (Applied Biosystems), according to the manufacturer’s instructions. All quantitative real-time RT-PCR reactions were assembled with TaqMan 2× PCR Master Mix and predeveloped gene expression probes from Applied Biosystems. Samples were analyzed on a CFX96 Real Time system using CFX manager software (Bio-Rad Laboratories). Gene expression levels were normalized to GAPDH or CD31, as indicated. All TaqMan probes were purchased from Applied Biosystems: human GAPDH (Hs99999905_m1), human VCAM-1 (Hs01003372_m1), human IRF-1 (Hs00971960_m1), mouse CD31 (Mm01242584_m1), and mouse VCAM-1 (Mm01320970_m1).
Immunofluorescence microscopy.
Serial 5-µm sections of artery were fixed in acetone for 10 min, air dried, rehydrated in PBS, and then stained with purified rat anti–mouse VCAM-1 (BioLegend) and an Alexa Fluor 594–conjugated goat anti–rat secondary antibody (Invitrogen). To visualize differences in VCAM-1 staining, the VCAM-1 antibody was titrated down to subsaturating levels. 5-µm sections of kidney were fixed as above and stained with purified rabbit anti–mouse CD31 (Abcam), rat anti–mouse CD31 (eBioscience), rabbit anti–mouse IRF-1 (Santa Cruz Biotechnology, Inc.), rat anti–mouse CD45 (BioLegend), or rat anti–mouse VCAM-1. Slides were then stained with Alexa Fluor 594–conjugated goat anti–rat and Alexa Fluor 488–conjugated donkey anti–rabbit secondary antibodies (Invitrogen). ProLong Gold antifade reagent with DAPI (Invitrogen) was applied to sections immediately before mounting and imaging. Immunofluorescence images were acquired with an Axiovert 200M fluorescence microscope (Carl Zeiss) equipped with an ORCA-AG digital camera (Hamamatsu Photonics) using Volocity software (PerkinElmer). A DAPI filter was used to detect nuclei, an FITC filter was used to detect Alexa Fluor 488–stained cells, and a tetramethylrhodamine isothiocyanate filter was used to detect Alexa Fluor 594–stained cells. To analyze renal tissue, the number of CD45+ leukocytes infiltrating into or immediately abutting individual glomeruli (identified by morphology under phase microscopy) were counted. 20–25 glomeruli were counted per mouse and averaged.
Statistical analysis.
Data are expressed as means ± SEM. Statistical comparisons were made using the Student’s t test. Comparisons between multiple groups were made using ANOVA with Tukey’s post-hoc test. In all experiments, unless otherwise stated, P ≤ 0.05 was considered statistically significant.
Acknowledgments
We thank L. Benson and G. Davis for assistance in EC culture and animal care.
This work is supported by National Institutes of Health (NIH) grant R01-HL109455 to J.S. Pober. C. Wang was supported by an NIH Medical Scientist Training Program grant (T32-GM007205) and is currently supported by an NIH National Research Service Award predoctoral fellowship (F30HL114253).
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- ChIP
- chromatin immunoprecipitation
- EC
- endothelial cell
- MFI
- median fluorescence intensity
- mTOR
- mammalian target of rapamycin
- rapa-EC
- rapamycin-treated EC
References
- Ahmad M., Marui N., Alexander R.W., Medford R.M. 1995. Cell type-specific transactivation of the VCAM-1 promoter through an NF-κB enhancer motif. J. Biol. Chem. 270:8976–8983 10.1074/jbc.270.15.8976 [DOI] [PubMed] [Google Scholar]
- Al-Lamki R.S., Bradley J.R., Pober J.S. 2008. Endothelial cells in allograft rejection. Transplantation. 86:1340–1348 10.1097/TP.0b013e3181891d8b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook-Mills J.M., Marchese M.E., Abdala-Valencia H. 2011. Vascular cell adhesion molecule-1 expression and signaling during disease: regulation by reactive oxygen species and antioxidants. Antioxid. Redox Signal. 15:1607–1638 10.1089/ars.2010.3522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritsche-Guenther R., Witzel F., Sieber A., Herr R., Schmidt N., Braun S., Brummer T., Sers C., Blüthgen N. 2011. Strong negative feedback from Erk to Raf confers robustness to MAPK signalling. Mol. Syst. Biol. 7:489 10.1038/msb.2011.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacinto E., Facchinetti V., Liu D., Soto N., Wei S., Jung S.Y., Huang Q., Qin J., Su B. 2006. SIN1/MIP1 maintains rictor-mTOR complex integrity and regulates Akt phosphorylation and substrate specificity. Cell. 127:125–137 10.1016/j.cell.2006.08.033 [DOI] [PubMed] [Google Scholar]
- Koch A.E., Burrows J.C., Haines G.K., Carlos T.M., Harlan J.M., Leibovich S.J. 1991. Immunolocalization of endothelial and leukocyte adhesion molecules in human rheumatoid and osteoarthritic synovial tissues. Lab. Invest. 64:313–320 [PubMed] [Google Scholar]
- Laplante M., Sabatini D.M. 2012. mTOR signaling in growth control and disease. Cell. 149:274–293 10.1016/j.cell.2012.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechleitner S., Gille J., Johnson D.R., Petzelbauer P. 1998. Interferon enhances tumor necrosis factor–induced vascular cell adhesion molecule 1 (CD106) expression in human endothelial cells by an interferon-related factor 1–dependent pathway. J. Exp. Med. 187:2023–2030 10.1084/jem.187.12.2023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madge L.A., Pober J.S. 2001. TNF signaling in vascular endothelial cells. Exp. Mol. Pathol. 70:317–325 10.1006/exmp.2001.2368 [DOI] [PubMed] [Google Scholar]
- Manes T.D., Pober J.S., Kluger M.S. 2006. Endothelial cell-T lymphocyte interactions: IP[corrected]-10 stimulates rapid transendothelial migration of human effort but not central memory CD4+ T cells. Requirements for shear stress and adhesion molecules. Transplantation. 82:S9–S14 10.1097/01.tp.0000231356.57576.82 [DOI] [PubMed] [Google Scholar]
- O’Brien K.D., Allen M.D., McDonald T.O., Chait A., Harlan J.M., Fishbein D., McCarty J., Ferguson M., Hudkins K., Benjamin C.D., et al. 1993. Vascular cell adhesion molecule-1 is expressed in human coronary atherosclerotic plaques. Implications for the mode of progression of advanced coronary atherosclerosis. J. Clin. Invest. 92:945–951 10.1172/JCI116670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pober J.S., Sessa W.C. 2007. Evolving functions of endothelial cells in inflammation. Nat. Rev. Immunol. 7:803–815 10.1038/nri2171 [DOI] [PubMed] [Google Scholar]
- Ren B., Deng Y., Mukhopadhyay A., Lanahan A.A., Zhuang Z.W., Moodie K.L., Mulligan-Kehoe M.J., Byzova T.V., Peterson R.T., Simons M. 2010. ERK1/2-Akt1 crosstalk regulates arteriogenesis in mice and zebrafish. J. Clin. Invest. 120:1217–1228 10.1172/JCI39837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts P.J., Der C.J. 2007. Targeting the Raf-MEK-ERK mitogen-activated protein kinase cascade for the treatment of cancer. Oncogene. 26:3291–3310 10.1038/sj.onc.1210422 [DOI] [PubMed] [Google Scholar]
- Sarbassov D.D., Ali S.M., Sengupta S., Sheen J.H., Hsu P.P., Bagley A.F., Markhard A.L., Sabatini D.M. 2006. Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol. Cell. 22:159–168 10.1016/j.molcel.2006.03.029 [DOI] [PubMed] [Google Scholar]
- Sun C., Alkhoury K., Wang Y.I., Foster G.A., Radecke C.E., Tam K., Edwards C.M., Facciotti M.T., Armstrong E.J., Knowlton A.A., et al. 2012. IRF-1 and miRNA126 modulate VCAM-1 expression in response to a high-fat meal. Circ. Res. 111:1054–1064 10.1161/CIRCRESAHA.112.270314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tellides G., Pober J.S. 2007. Interferon-gamma axis in graft arteriosclerosis. Circ. Res. 100:622–632 10.1161/01.RES.0000258861.72279.29 [DOI] [PubMed] [Google Scholar]
- Tsoyi K., Jang H.J., Nizamutdinova I.T., Park K., Kim Y.M., Kim H.J., Seo H.G., Lee J.H., Chang K.C. 2010. PTEN differentially regulates expressions of ICAM-1 and VCAM-1 through PI3K/Akt/GSK-3β/GATA-6 signaling pathways in TNF-α-activated human endothelial cells. Atherosclerosis. 213:115–121 10.1016/j.atherosclerosis.2010.07.061 [DOI] [PubMed] [Google Scholar]
- Wang C., Yi T., Qin L., Maldonado R.A., von Andrian U.H., Kulkarni S., Tellides G., Pober J.S. 2013. Rapamycin-treated human endothelial cells preferentially activate allogeneic regulatory T cells. J. Clin. Invest. 123:1677–1693 10.1172/JCI66204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X.S., Vanderziel C., Bennett C.F., Monia B.P. 1998. A role for c-Raf kinase and Ha-Ras in cytokine-mediated induction of cell adhesion molecules. J. Biol. Chem. 273:33230–33238 10.1074/jbc.273.50.33230 [DOI] [PubMed] [Google Scholar]