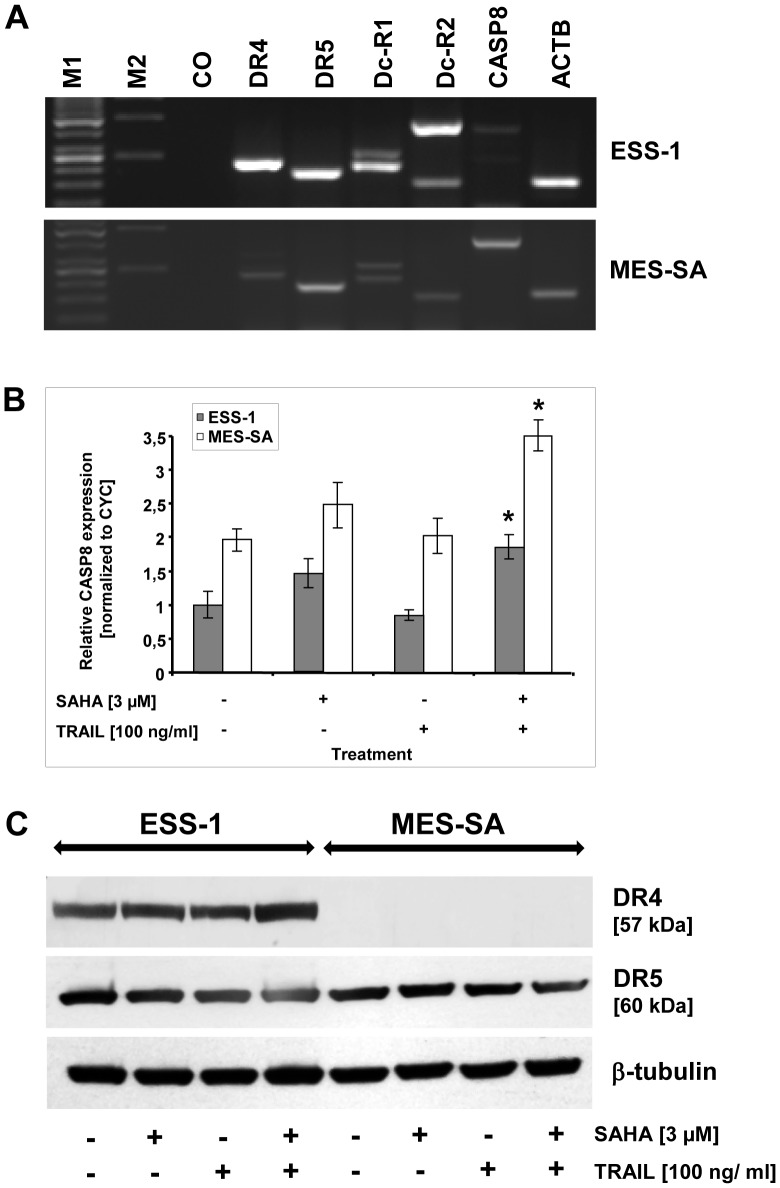
Figure 4. Reduced expression of caspase-8 in ESS-1 cells and DR4 (TRAIL-R1) in MES-SA cells.
TRAIL receptors (TRAIL-R1/DR4 and TRAIL-R2/DR5), both TRAIL decoy receptors (Dc-R1, Dc-R2), and caspase-8 (CASP8) were amplified from cDNA in order to monitor defects in gene expression (A). Therefore, RNA of untreated cells was isolated, reversely transcribed, and subjected to qRT-PCR with primers binding to exonic sequences. PCR products were run on a 1.5% agarose gel, stained with ethidium bromide, and photographed. Amplification of beta-actin with or without genomic DNA served as a positive control (ACTB) or negative control (CO), respectively. Note the weaker bands for caspase-8 and DR4 in ESS-1 and MES-SA cells, respectively. The additional high molecular weight band for Dc-R2 in ESS-1 cells represents the genomic amplicon. M1, Gene ruler 50 bp DNA ladder; M2, λBst91I marker. (B) The relative caspase-8 expression of SAHA and/or TRAIL treated ESS-1, and MES-SA cells, was quantitated by qRT-PCR. Technical triplicate cycle threshold (ct) values of three biological replicates were normalized to cyclophillin (CYC) expression. Asterisks (* p<0.05) indicate statistically significant differences as compared to the untreated control. (C) Western blot analysis of ESS-1 and MES-SA cells treated for 8 hours with 3 μM SAHA and/or 100 ng/ml TRAIL. Untreated cells were used as control. Cell extracts were prepared, then 30 μg of protein subjected to SDS-PAGE (4 to 12% Bis-tris gel), and immunoblotted with the indicated antibodies against DR4 (TRAIL-R1), DR5 (TRAIL-R2) and β-tubulin as loading control. The molecular weights of presented bands are indicated in brackets.