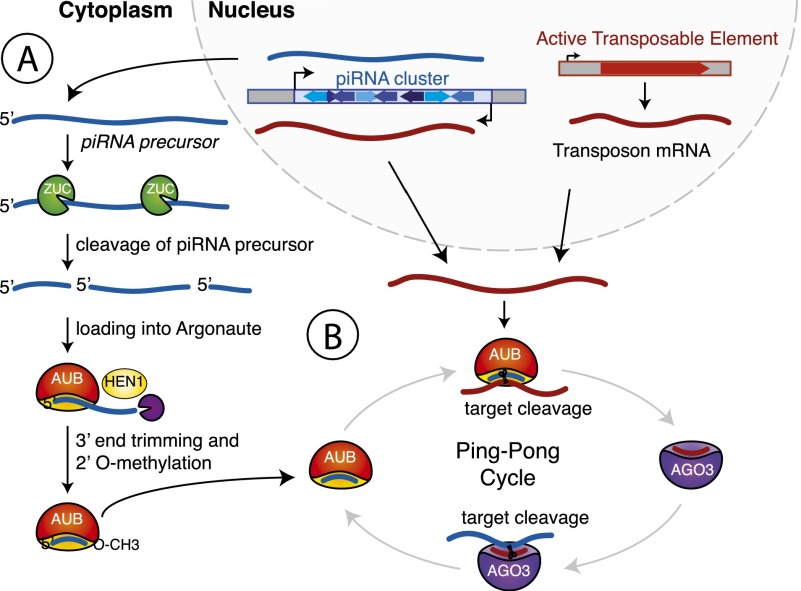
Figure 2.
Biogenesis of piRNAs. (A) Long RNAs that constitute substrates for piRNA processing are transcribed from distinct genomic regions called piRNA clusters. These transcripts are cleaved, presumably by the endonuclease Zucchini (ZUC), to produce 5′ phosphorylated piRNA precursors. The precursors are anchored into Piwi proteins with their 5′ end, and their 3′ end is trimmed by an unknown nuclease. Methylation of the 3′ end 2′OH group by the methyltransferase HEN1 produces mature piRNAs. (B) The Ping-Pong cycle amplifies piRNA sequences. piRNA-loaded Aubergine (AUB) initiates the Ping-Pong cycle by cleaving a complementary target transcript derived from either an active TE or the opposite strand of the initial cluster. This produces the 5′ end of a new piRNA precursor. This precursor is loaded into Argonaute 3 (AGO3) and guides cleavage of a piRNA cluster transcript complementary to its sequence.