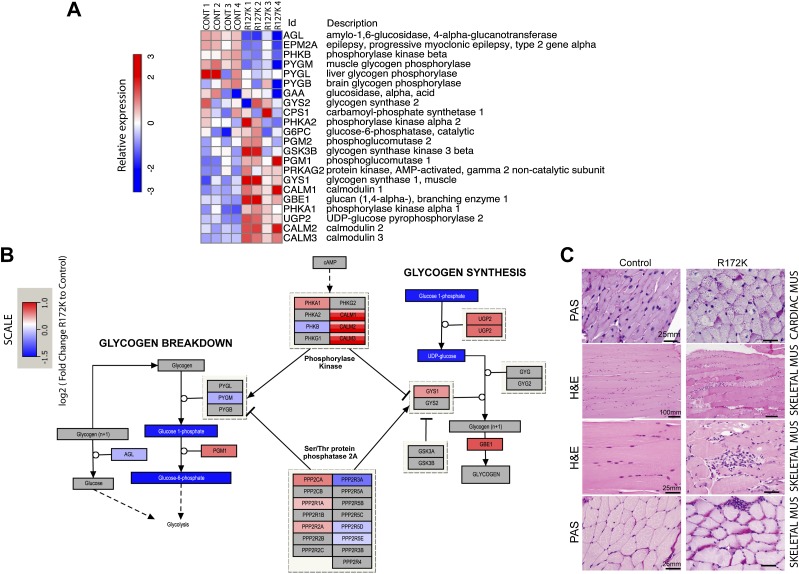
Figure 5.
Transcriptional and metabolic changes in IDH2 mutant hearts compared with controls. (A) Expression of the genes involved in glycogen biosynthesis and degradation in R172K mutant hearts compared with controls. (B) Pathway map of enzymes, metabolites, and polysaccharides in the glycogen biosynthesis pathway. Color scales indicating RNA levels of the enzymes are the same as in A. The color scale for metabolites indicates log2 fold change of levels in R172K mutant hearts (n = 9) relative to controls (n = 10). Metabolites that were not measured or had no detectable change are shown in gray. P-values for UDP glucose and G1P are P < 0.05 and P < 0.007, respectively. (C) Histology of the cardiac and skeletal muscles. PAS-stained sections of cardiac muscle showing increased collections of diastase-sensitive glycogen in R172K animals compared with control. H&E-stained sections of skeletal muscle show myopathic changes, including increased variation in fiber size, increased numbers of internalized nuclei, and increased endomysial fibrosis. Myofiber regeneration, degeneration, and necrosis are present in association with multifocal myophagocytosis. PAS stains highlight increased glycogen deposition in IDH2 mutant skeletal muscles compared with control. Bars show the indicated lengths and are the same across images on the same row.