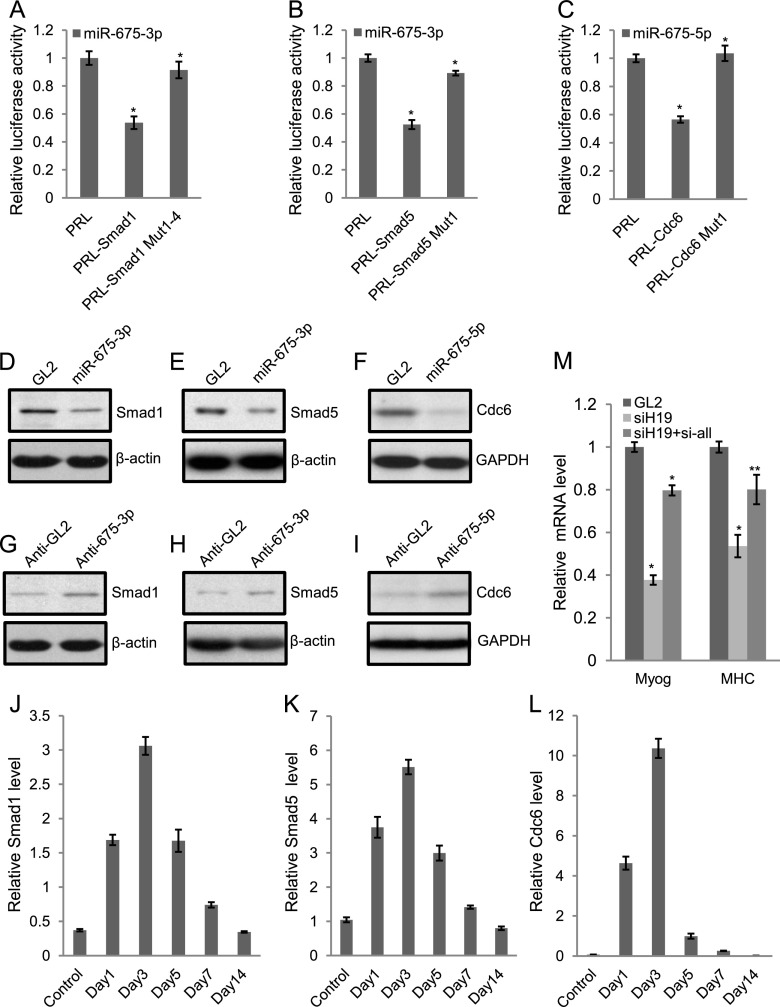
Figure 5.
Smad1, Smad5, and Cdc6 are important targets for miR-675-3p and miR-675-5p. (A–C) Cotransfection of miR-675-3p or miR-675-5p microRNA into U2OS osteosarcoma cells repressed Renilla (rr) luciferase activity when the rr reporter was fused to the target 3′ UTR. Mutations in the target sites in the 3′ UTR (Supplemental Fig. 7) relieved the repression. A firefly (pp) luciferase plasmid was cotransfected as a transfection control. The rr/pp was normalized to that for a control Renilla luciferase plasmid with a vector 3′ UTR (PRL) and expressed relative to the normalized rr/pp in cells transfected with the GL2 RNA control. Mean ± SEM of three individual experiments. (*) P < 0.001. (D–F) Transfection of miR-675-3p or miR-675-5p in C2C12 myoblast cells in GM decreased Smad1, Smad5, and Cdc6 protein levels as detected by Western blotting 2 d after the second transfection. GL2 served as a control oligonucleotide. β-Actin or GAPDH was used as a loading control. (G–I) 2′O-methyl oligonucleotide against miR-675-3p or LNA antisense oligonucleotide against miR-675-5p (anti-675-3p or anti--675-5p) increased endogenous Smad1, Smad5, and Cdc6 protein levels in C2C12 cells cultured in DM for 2 d. 2′O-methyl or LNA antisense GL2 (anti-GL2) served as a control oligonucleotide. β-Actin or GAPDH is used as a loading control. (J–L) As measured by qRT–PCR, miR-675-3p or miR-675-5p target genes display anti-correlation in their temporal expression pattern during regeneration of skeletal muscle after CTX injury. (M) C2C12 myoblast cells were transfected with GL2 control siRNA, H19 siRNA, or H19 siRNA combined with siRNAs specific to Smad1, Smad5, and Cdc6 once in GM and then on DM1, and cells were harvested on DM3. qRT–PCR was performed for myogenin and MHC. The normalization for myogenin and MHC mRNAs was as described in Figure 1. Mean ± SEM of three biological replicates are shown. (*) P < 0.001; (**) P < 0.005.