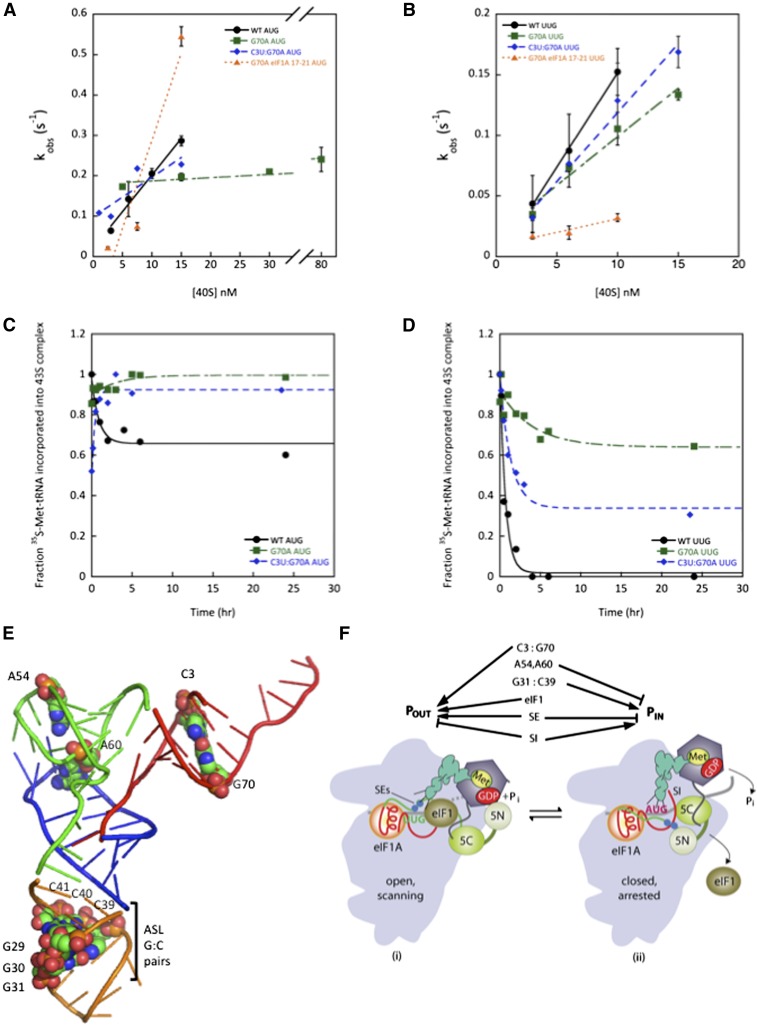
Figure 6.
Disrupting acceptor stem base pair C3:G70 shifts the equilibrium from POUT to PIN. (A,B) Determination of kon values for TC association with 40S•eIF1•eIF1A complexes and mRNA(AUG) (A) or mRNA(UUG) (B). Each value is the average of at least two independent experiments. Errors are average deviations. (C,D) Analysis of TC dissociation from 43S·mRNA complexes for mRNA(AUG) (C) or mRNA(UUG) (D) complexes. Representative curves from at least two independent experiments are shown. koff values and endpoints for dissociable complexes are given in Supplemental Table S2. (E) PyMol rendering of the crystal structure of yeast tRNAi (Protein Data Bank [PDB]: 1YFG) using color-coding to designate the acceptor stem (red), T-stem–loop (green), D-stem–loop (blue), and ASL (gold) and depicting by spheres the bases or base pairs implicated here in start codon recognition. (F) Model summarizing the deduced roles of conserved tRNAi residues in start codon recognition. See Supplemental Figure S2 for description of the open/POUT and closed/PIN states of the PIC and roles of eIF1 and the SEs/SI elements of eIF1A in regulating conformational rearrangements and reactions accompanying AUG recognition. Results in this study indicate that base pair C3:G70 functions together with eIF1 and eIF1A SEs to stabilize the POUT conformation of TC binding, whereas residues A54/A60 impede rearrangement to the PIN state in a manner overcome efficiently only with the perfect codon:anticodon duplex formed at AUG. G31:C39 and most likely the other two ASL G:C pairs are required for thermodynamic coupling between AUG and tRNAi in the PIN state. Not summarized here is the fact that replacing G31:C39 with other Watson-Crick pairs further stabilizes PIN and thereby increases initiation at NUG near-cognates (see Supplemental Figs. S11–S13 for further details.)