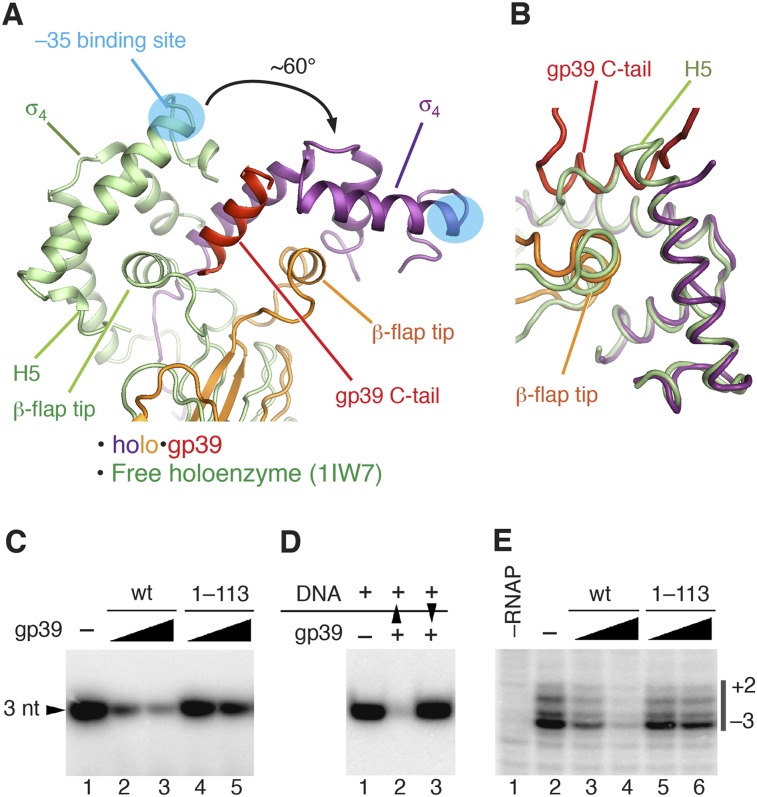
Figure 4.
The mechanism of transcription inhibition by gp39. (A,B) Superimposition of holo•gp39 and the free holoenzyme (Protein Data Bank [PDB] 1IW7; green) (Vassylyev et al. 2002). (A) The core modules of the two RNAP structures were superimposed by minimizing the root mean square deviation (RMSD) between the Cα atoms. (B) Superimposition of the σ4 domains of the two structures. (C) Transcription inhibition by wild-type gp39 (wt) and its truncated variant lacking the C-terminal tail (1–113) on a −10/−35 promoter (T5 N25). (D) Analysis of transcription inhibition by gp39 added either before (lane 2) or after (lane 3) the promoter DNA. Black triangles indicate order of addition. (E) Analysis of gp39 (wild type [wt] and 1–113) on promoter melting by potassium permanganate probing. Thymine positions in the melted promoter region (numbered relative to the transcription starting point) are indicated on the right.