Figure 7.
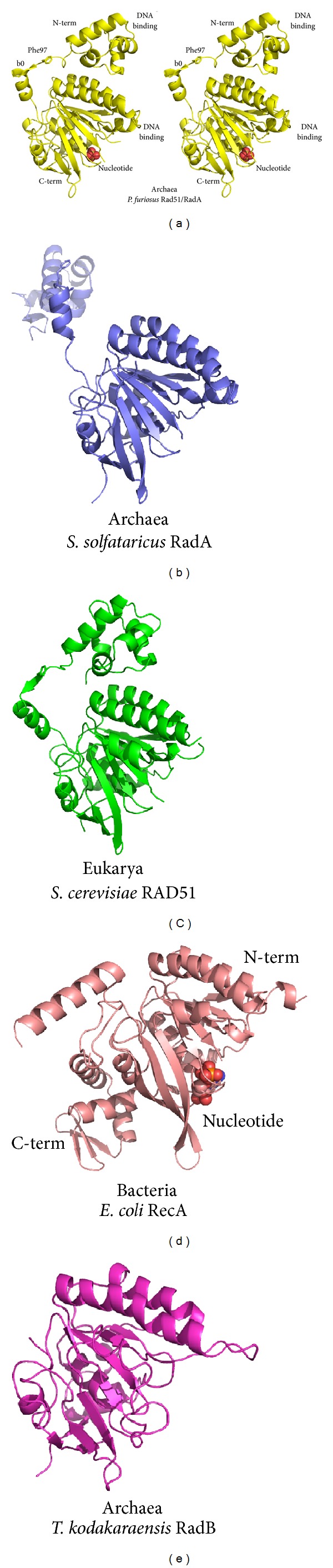
Structural comparisons of Rad51 family proteins. (a) Stereoview of archaeal P. furiosus Rad51 (PDB code 1PZN). While Rad51 generally forms larger homopolymeric assemblies, the prototypical fold of a single archaeal Rad51 or RadA protein consists of a small 4-helix bundle N-terminal DNA-binding domain, which is tethered to a larger C-terminal ATPase domain. The ATPase domain has several loops that are also implicated in binding DNA. The interdomain linker that tethers the two domains contains a polymerization motif (PM) that consists of a β-strand β0, which, upon contact with a neighboring subunit, extends the β-sheet of the ATPase domain. Conserved Phe97 forms a ball and socket to stabilize this interaction. (b) The two domains of the archaeal S. solfataricus structure individually superpose well with the PfRad51 domains. However, this structure reveals the flexibility of the interdomain linker, where the N-terminal domain has swung outward (PDB code 2BKE). (c) The structure of S. cerevisiae RAD51 reveals structural conservation with the archaeal proteins (PDB code 1SZP). (d) The bacterial RecA protein shares the ATPase domain fold (PDB code 2REB). However, in this structure the ATPase is represented in the N-terminal domain, while in archaea and eukarya the ATPase is represented in the C-terminal domain. A small N-terminal arm extends from the bacterial ATPase and serves a similar function as the Rad51 PM. (e) The archaeal T. kodakaraensis RadB protein structure illuminates how extensions to the ATPase likely served as critical components of primordial recombination structures, where in the archaea and eukarya the N-terminal domain became an accessory domain, whereas in bacteria the C-terminus gave rise to an accessory domain (PDB code 2CVF).
