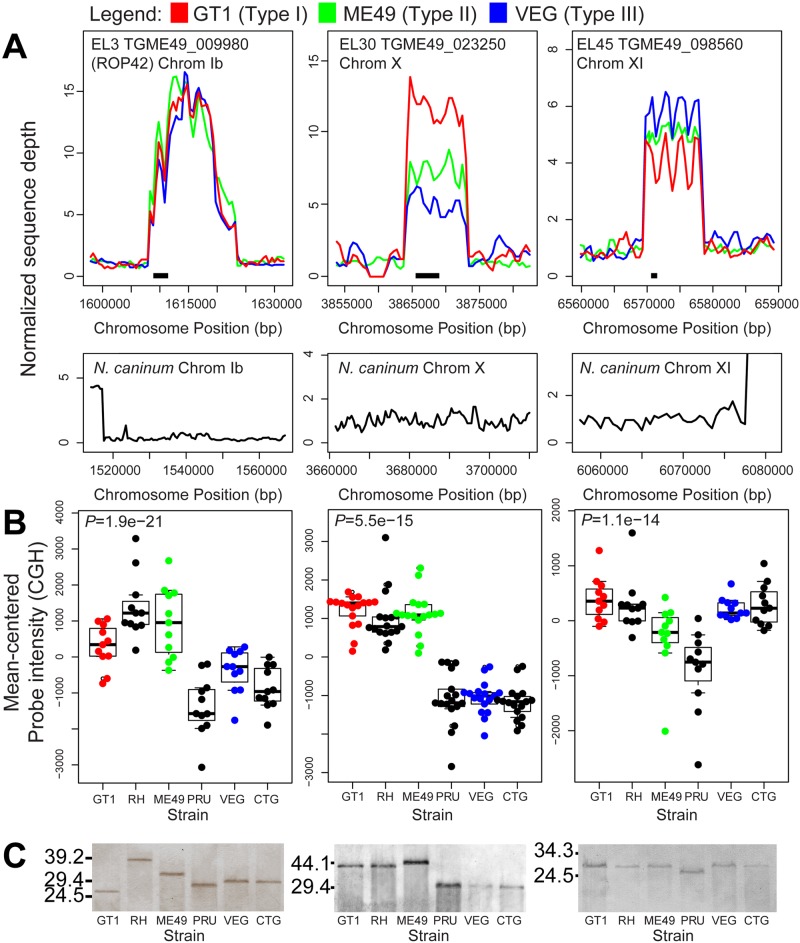
FIG 5 .
Sequence read, comparative genomic hybridization, and Southern blot analysis for EL3, EL30, and EL45. (A) Sequence read analysis for three strains of T. gondii (top) and N. caninum Liverpool (bottom). Data for each strain and locus are normalized to the read coverage in the 20 kb flanking the expanded locus to the left. (B) Comparative genomic hybridization (CGH) across 6 T. gondii strains, 2 from each of the 3 canonical lineages. Only microarray probes with perfect matches in GT1, ME49, and VEG were used in the calculations. Boxes span the first and third quartiles and contain the median value for all useful probes. P values for significant differences in hybridization intensity compared to the single-copy gene AMA1 (gray horizontal line) are at the top of the graph, and individual values are shown in the bee swarm plots. (C) Southern blots for 6 T. gondii strains using DNA digested with restriction enzymes predicted to cut outside the repeat locus. Restriction enzymes were BspEI, BglI, and NotI, respectively. A similar blot for the single-copy gene AMA1 can be found in the supplemental material. Numbers at left are molecular masses in kilobases.