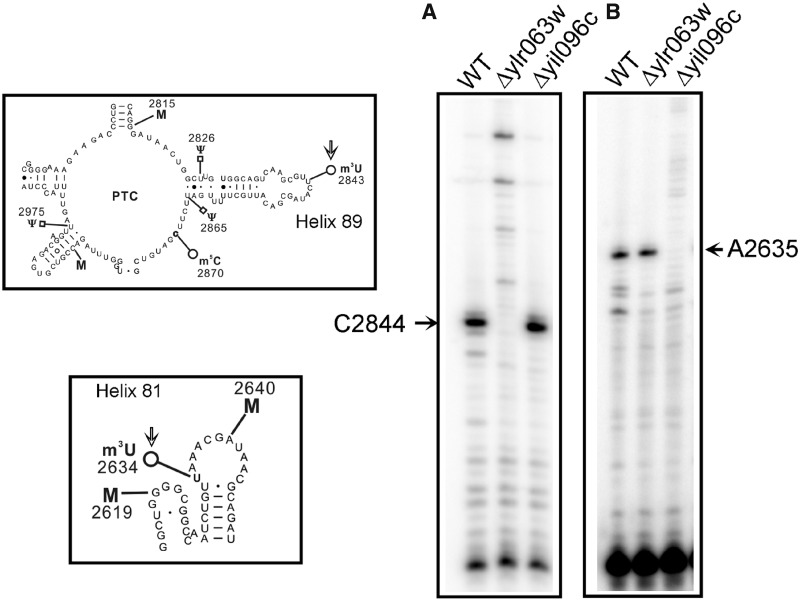
Figure 4.
Primer extension analysis. To further substantiate the role of Yil096c and Ylr063w in the methylation of m3U residues and validate the positions of the m3U modifications in the 25S rRNA, we performed the primer extension analysis with the 25S rRNA isolated from Δyil096c and Δylr063w deletion mutants along with isogenic wild type. The methylation of the uridine at N3 blocks the Watson–Crick base pairing, and results in a strong stop in the primer extension analysis (A) The presence of m3U at position 2843 in the 25S rRNA from the wild type and Δyil096c cells led to a strong stop at position 2844. However, this stop was absent in Δylr063w deletion mutant. (B) Similarly, owing to presence of an m3U residue at position 2634, a strong stop at position 2635 was observed in wild type and Δylr063w but was missing in Δyil096c deletion mutant. The positions of the methylated residues were determined accurately from running side by side a sequencing ladder (data not shown), is given on the sides of the gels, and the bands of interest are shown with arrows. This clearly validated the specific involvement of Ylr063w and Yil096c in the methylation of m3U2843 and m3U2634, respectively. Both helices 81 and 89, carrying m3U residues (marked with an arrow) are displayed on left of the panel A and B.