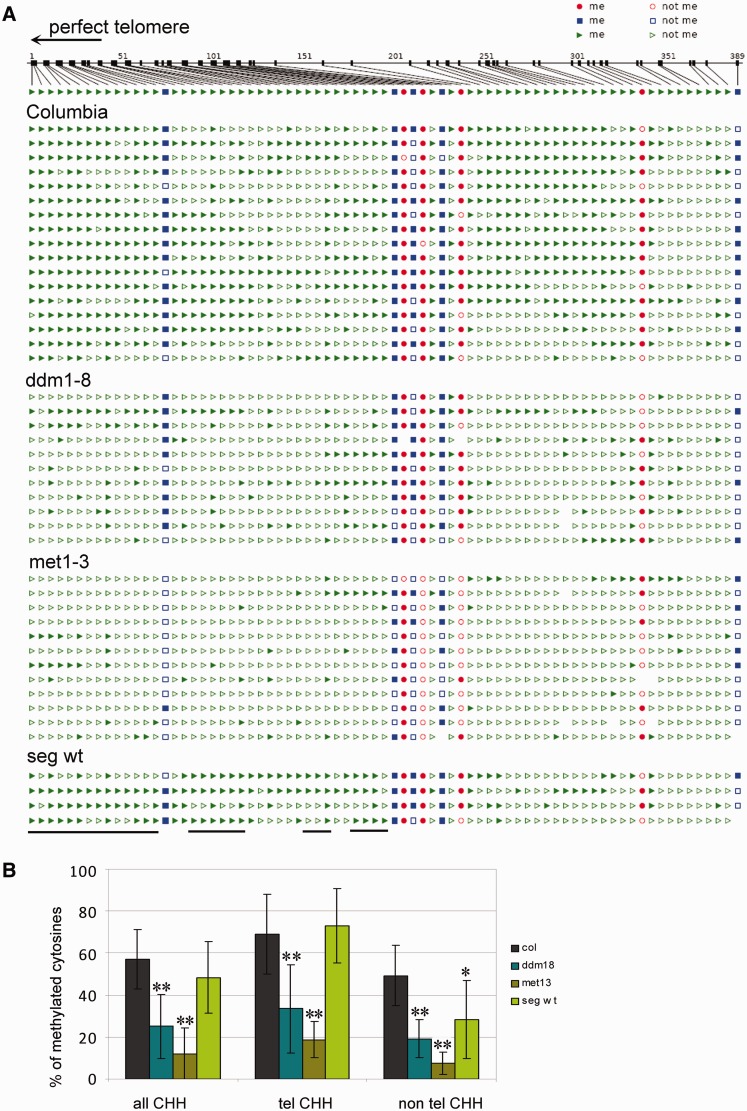
Figure 3.
Analysis of cytosine methylation in the proximal part of the 1L telomere arm. (A) Distribution of methylated cytosines along the 389-bp region, which is delimited by primers derived from the subtelomeric region and a specific insertion into the 1L telomere (8). Seventeen clones from three Columbia plants, 11 clones from two ddm1-8 G2 plants, 12 clones from two met1-3 G2 plants and 4 clones from a segregated wt plant (seg wt) were analysed. Red circles, CG methylation; blue squares, CHG methylation; green triangles, CHH methylation; filled symbols, methylated cytosine; empty symbols, non-methylated cytosine. Positions of cytosines located in perfect telomeric repeats are delimited by the black lines below the figure. The arrowhead determines the direction to the perfect telomere. (B) Graphical representation of the level of all methylated cytosines located in non-asymmetrical sequences (all CHH), in perfect telomeric repeats (tel CHH) and outside these repeats (non-tel CHH). Standard deviations reflect variability between clones. The level of methylated cytosines is significantly lower in methylation mutant clones (**P < 0.01 for all sequence contexts). In the segregated wt, methylation of cytosines outside of telomeric repeats (non-tel CHH) is reduced (*P < 0.05).