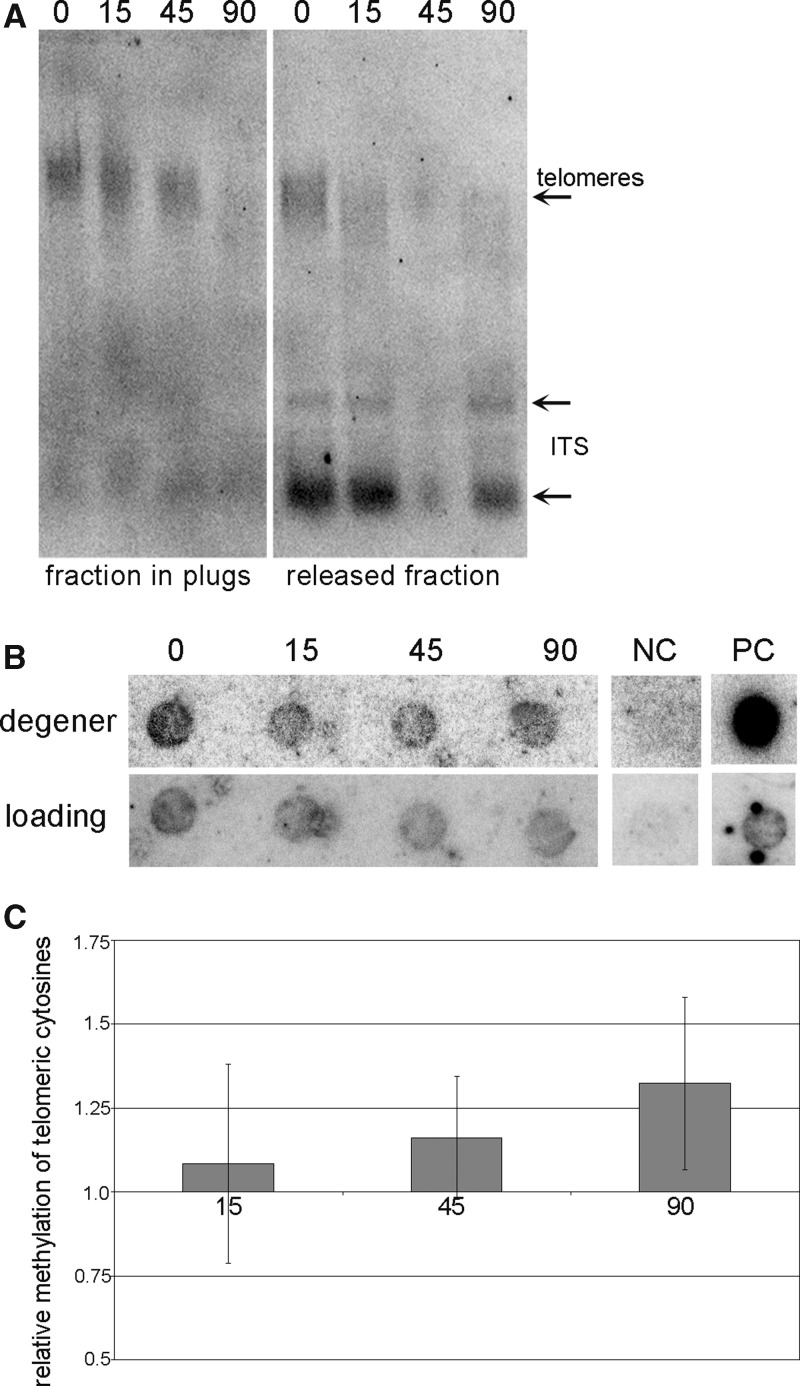
Figure 4.
Analysis of relative methylation of cytosines in telomeric repeats. (A) High-molecular weight DNA in agarose plugs was digested by Bal31 exonuclease and analysed by TRF. The fraction retained in agarose plugs and the fraction released into the solution were analysed separately. The time of Bal31 digestion (in min) is marked above the lines. The positions of telomeres and ITSs are depicted by arrowheads. (B) DNA isolated from agarose plugs was modified by sodium bisulfite and analysed by Southern hybridization with radioactively labelled ‘loading’ and ‘degener’ probes. The time of BAL31 digestion (in min) is depicted. NC, negative control, DNA from the plasmid pUC19; PC, positive control, non-modified genomic DNA isolated from A. thaliana leaves. (C) Relative methylation of telomeric cytosines in Bal31-digested samples. Hybridization signals were evaluated by the MultiGauge software (FujiFilm), and expressed as the ‘degener’/‘loading’ ratio. The ratio in BAL31 non-digested samples was arbitrary taken as 1. Five independent experiments were evaluated.