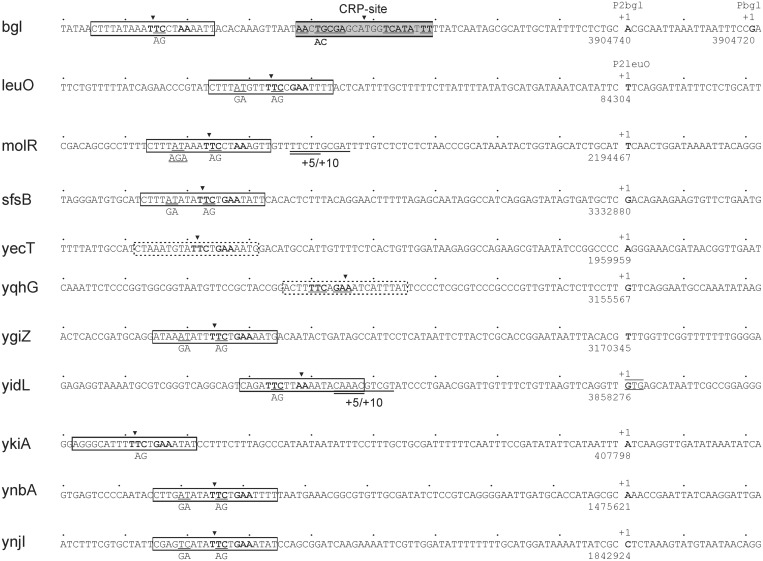
Figure 2.
Sequence of BglJ–RcsB activated promoters and their BglJ–RcsB binding sites. The transcription starts sites (+1) were mapped by 5′-RACE of RNA isolated from strain T1166 constitutively expressing BglJ, as well as of RNA isolated from strain T75 carrying a bglJ deletion (allele ΔyjjP-yjjQ-bglJ). The positions of the transcription start sites refer to the MG1655 genome sequence NC_000913. For 5′-RACE analysis of bgl also RNA of the Δhns stpA ΔyjjP-yjjQ-bglJ mutant strain T1048 was used. This revealed the transcription start site mapping at position 3904720 in the bglJ deletion background (T75, ΔyjjP-yjjQ-bglJ) and Δhns stpA ΔyjjP-yjjQ-bglJ (T1048), i.e. the absence of BglJ, while the transcription start site at position 3904740 is used in the BglJ positive strain T1166. The BglJ–RcsB activated leuO P2 promoter was mapped previously (20). BglJ–RcsB binding sites marked by boxes with solid lines were mapped by mutagenesis in this work (molR, sfsB, yecT, ygiZ, yidL, ykiA, ynbA and ynjI) or previously [bgl, leuO (4,20)]. Site-specific mutations are indicated underneath the sequence. Insertions of 5 and 10 bp were generated by duplication of the underlined sequences (+5/+10). Predicted BglJ–RcsB binding sites are marked by boxes with dashed lines (yecT and yqhG). The transcription start site of the yidL promoter maps to the first base of a GTG codon (underlined) that is annotated as translation start site (33).