Abstract
NKAP is a highly conserved protein with roles in transcriptional repression, T-cell development, maturation and acquisition of functional competency and maintenance and survival of adult hematopoietic stem cells. Here we report the novel role of NKAP in splicing. With NKAP-specific antibodies we found that NKAP localizes to nuclear speckles. NKAP has an RS motif at the N-terminus followed by a highly basic domain and a DUF 926 domain at the C-terminal region. Deletion analysis showed that the basic domain is important for speckle localization. In pull-down experiments, we identified RNA-binding proteins, RNA helicases and splicing factors as interaction partners of NKAP, among them FUS/TLS. The FUS/TLS–NKAP interaction takes place through the RS domain of NKAP and the RGG1 and RGG3 domains of FUS/TLS. We analyzed the ability of NKAP to interact with RNA using in vitro splicing assays and found that NKAP bound both spliced messenger RNA (mRNA) and unspliced pre-mRNA. Genome-wide analysis using crosslinking and immunoprecipitation-seq revealed NKAP association with U1, U4 and U5 small nuclear RNA, and we also demonstrated that knockdown of NKAP led to an increase in pre-mRNA percentage. Our results reveal NKAP as nuclear speckle protein with roles in RNA splicing and processing.
INTRODUCTION
Within the nucleus, nuclear speckles are one of the most prominent nuclear compartments (1,2). They were initially described by Cajal using silver staining procedures as pale-rounded or irregular areas of homogeneous texture in the neuronal nucleus (3). Nuclear speckles are dynamic structures involved in the storage, assembly and recycling of pre-messenger RNA (mRNA) processing factors and in the post-transcriptional processing of pre-mRNAs (4–6). Nuclear speckles become round and increase in size on transcriptional inhibition, which supports the view that speckles might function in the storage, assembly and modification of splicing factors (5–8). They steadily exchange their components with the nucleoplasm and active transcription factories, thereby facilitating the expression of multiple active genes, some of which associate directly with the edge of nuclear speckles (9).
Nuclear speckles or their ultrastructural equivalent, the interchromatin granule clusters, is enriched in components of the pre-mRNA splicing machinery, particularly spliceosomal small nuclear ribonucleoprotein particles (snRNPs) and other non-snRNP protein splicing factors such as SRSF2 and U2AF (1,4,10). They also contain several kinases, the PP1 phosphatase, 5′- and 3′-end processing factors, some transcription factors involved in gene expression driven by RNA polymerase II and a population of poly(A) RNA and non-coding RNA (ncRNA) (1,2,4). For localization to nuclear speckles, the arginine/serine-rich domain (RS domain) has been shown to be necessary and sufficient, and the proteins are known as SR proteins (11–13).
In humans, the SR protein family has a common structural organization containing either one or two RNA recognition motifs (RRM) that provide RNA-binding specificity, and a variable-length RS domain that functions as a protein interaction domain (14). SR-like or SR-related proteins exhibit differences in domain structure and lack an RRM. Nevertheless, they have been shown to be involved in RNA splicing and metabolism (15). The RS domain, which is extensively phosphorylated, promotes protein–protein interactions that are essential for the recruitment of the splicing apparatus and for splice site pairing (16). RS domains have also been shown to directly contact the pre-mRNA branch point; thus, RS domains may not solely function through protein–protein interactions (17). In fact, in vivo depletion of the SR proteins SRSF2 and SRSF1 dramatically attenuated the production of nascent RNA, and SR or SR-related proteins are now generally accepted as important regulators of constitutive and alternative splicing of pre-mRNA (18).
In mammals, the DUF 926 domain is shared between two proteins, namely, NKAP (NF kappa B activating protein) and NKAP-L (NF kappa B activating protein-like). The name NKAP is derived from the previous findings that this protein could activate the NF kappa B pathway (19). More recently, it was reported that NKAP regulates the Notch signaling pathway and is required for T-cell development (20). This is achieved by interacting with HDAC3 and CBF1 interacting co-repressor. Receptor interacting protein was also shown as potential interaction partner for NKAP through yeast two-hybrid screening (19). It is also required for the maintenance and survival of hematopoietic stem cells (21). In regard to T-cell maturation, NKAP has an important role in the functional competency and maturation of T cells (22). These studies were carried out in mice with T-cell-specific ablation of the gene (20). Additionally, a role of NKAP is predicted in splicing and RNA metabolism, as it has been found as interacting partner of RNA binding proteins and as a known core component of splicing complexes through extensive yeast two-hybrid screening (23,24).
We have characterized and cloned NKAP as a SR-related protein that contains a domain rich in arginines and serines and a domain of unknown function, DUF 926, but lacks a recognizable RRM. We show that NKAP localizes to nuclear speckles and interacts with other RNA binding proteins such as heterogeneous nuclear ribonucleoproteins (hnRNPs), splicing factors and RNA helicases. Its localization during mitosis points out that the behavior of NKAP is similar to the one of other SR proteins. We also identified NKAP binding RNA segments in HEK 293T cells using crosslinking and immunoprecipitation (CLIP) followed by RNA-seq. Our analysis revealed global profiles of NKAP-mediated regulation of pre-mRNA splicing and association with splicing small nuclear RNAs (snRNAs).
MATERIALS AND METHODS
Cloning of murine NKAP and FUS/TLS complementary DNA, expression of recombinant protein and production of antibodies
NKAP and FUS/TLS complementary DNA (cDNA) were amplified from mRNA isolated from mouse brain. Full-length NKAP and polypeptides encompassing the N- and the C-terminal domains were cloned in pEGFP-C2 vector. The full-length FUS was cloned in pCMV-3Tag-6 and FUS polypeptides were cloned into pEGFP-C2 vector. The sets of primers used for cloning are mentioned in Supplementary Table S2. For the generation of NKAP-specific monoclonal antibodies, a C-terminal GST-tagged polypeptide encompassing the DUF 926 domain (residues 273–415) was used. Expression was in Escherichia coli BL21, and induction was with isopropyl β-D-1-thiogalactopyranoside (1 mM). The recombinantly expressed GST-tagged protein was purified using affinity chromatography with glutathione-Sepharose beads, and 50 µg of protein was used to immunize four female BALB/c mice, followed by four boosts of 50 µg each over 4 weeks. Hybridoma cells were generated. mAb K85-80-5 was used in this study.
Mammalian cell culture
HeLa and HEK293T cells were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum, penicillin and streptomycin (100 μg/ml), l-glutamine and non-essential amino acids. WI-38, a human fetal fibroblast cell culture line, was maintained in MEM supplemented with l-glutamine, 15% fetal bovine serum, non-essential amino acids, penicillin and streptomycin (100 μg/ml). For transfection of HEK293T cells, the calcium phosphate method was used. At 24 h after transfection, cells were collected for RNA binding assays. Transfection of HeLa cells was performed by Nucleofector Kit R (Lonza, Cologne, Germany) according to the manufacturer’s instructions. We designed two different small hairpin RNAs against human NKAP. The targeted sequences are mentioned in Supplementary Table S2. They were cloned into a pSHAG-1 vector and transfected in HEK293T cells using lipofectamine 2000 (Invitrogen).
Tissue preparation and western blotting
Mouse tissues were prepared as described previously (25). For analysis of tissue by immunoblotting, samples were frozen in liquid nitrogen and total protein extracts were prepared in 10 mM Tris-HCl, pH 7.8, 1 mM ethylene glycol tetraacetic acid, 1 mM DTT and 0.5 mM PMSF containing complete mini protease inhibitor cocktail. After pelleting of the cell debris, supernatants were subjected to standard sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) (12% acrylamide), and proteins were blotted to nitrocellulose membrane.
Actinomycin D and nocodazole treatment and immunofluorescence
HeLa cells were transfected with GFP-NKAP, GFP-RS+Basic and GFP-DUF plasmids and cultured overnight on glass coverslips in 24-well plates in growth medium. Actinomycin D (0.5 µg/ml; Sigma) was added, and cells were cultured at 37°C for 3 h. The cells were washed twice with PBS and subjected to immunofluorescence. For cell cycle synchronization, cells were treated with nocodazole (10 µM/ml for 3 h).
Mouse monoclonal antibodies for SRSF2 (Sigma, 1:1000) were used, and secondary antibodies for indirect immunofluorescence analysis were conjugated with Alexa 568 (Molecular Probes). Nuclei were visualized with the DNA-specific dye 4′,6-diamidino-2-phenylindole (Sigma). Fixation was done with 4% paraformaldehyde followed by Triton X-100 (0.5%) treatment. Specimens were analyzed by wide-field fluorescence microscopy (DMR, Leica) or confocal laser scanning microscopy (TCS-SP5, Leica). Image processing was done with Adobe Photoshop or the TCSNT software, respectively.
Protein–protein interaction studies
Interaction studies were done by immunoprecipitation of GFP-NKAP using GFP polyclonal antibodies from cell-free extracts of HeLa cells expressing GFP-NKAP. HeLa cells expressing GFP-NKAP were washed in 1× PBS and lysed in ice-cold lysis buffer (50 mM Tris-HCl, pH 8.0, 5 mM EDTA, 5 mM ethylene glycol tetraacetic acid, 10 mM MgCl2, 100 μM PMSF, 10 mM Na2H2P2O7, 1 mM ATP, 20 mM NaF, 1 mM Na3VO4, 1% PEG 8000, 1% Triton X-100, 150 μM beta mercaptoethanol and protease inhibitor cocktail) for 20 min on ice. The cleared lysate was incubated with protein A-Sepharose beads (GE Healthcare) for 1 h at 4°C on a rotary wheel for pre-clearing. After 1 h, the beads were removed by centrifugation, and the supernatant was incubated with protein A-Sepharose beads bound to GFP-specific polyclonal antibodies for 2 h. Afterward, beads were washed three times with lysis buffer without NaF, Na3VO4, ATP and Triton X-100. Pull-down eluates were resolved by SDS-PAGE (12% acrylamide), and bands were analyzed by liquid chromatography - mass spectrometry (LC-MS) (Zentrale Bioanalytik of the CMMC).
The interaction between NKAP and FUS was studied through GST pull-down assays. GST-RS+ Basic, GST-RS, GST-Basic and GST-DUF 926 were produced in E. coli. Cells were lysed and GST-fusion proteins were bound to glutathione-Sepharose 4B beads (GE Healthcare). Beads containing GST or beads only were used as a control. Lysates of HeLa cells expressing FLAG-FUS, GFP-QGSY, GFP-G Rich+RGG1, GFP-RGG2+ZnF+RGG3, GFP-RGG1, GFP-RGG2 and GFP-RGG3 were incubated with beads coupled with GST and GST fusion proteins for 2 h. Afterward, beads were washed three times with washing buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl). Pull-down eluates were resolved by SDS-PAGE (12% acrylamide) and immunoblotted with polyclonal FLAG antibodies (Sigma).
RNA binding assays
Capped transcripts were generated as described previously (26). In vitro splicing reactions were performed for 2 h in HeLa cell nuclear extract (CIL Biotech) that was supplemented with HEK293 whole-cell extracts expressing the indicated FLAG-tagged proteins (26). HEK293 whole-cell extracts and FLAG-immunoprecipitations were performed with anti-FLAG agarose beads (Sigma) as previously described (26). RNAs were recovered by TRIzol extraction and isopropanol precipitation and analyzed by denaturing PAGE. Ten percent of the total input material was loaded in the input panels.
RNA binding assay-GST tagged RS+Basic, RS, Basic and DUF926 were expressed in E. coli BL21 cells and purified in the presence of protease inhibitor mixture (Sigma). Poly(U)- and poly(C)-Sepharose 4B (Sigma) were equilibrated in washing buffer. The purified proteins were incubated with poly(U)- and poly(C)-Sepharose 4B, respectively, at 4°C for 1 h with shaking. The beads were washed extensively with washing buffer, and the bound protein was resuspended in SDS sample buffer. The supernatant was analyzed by SDS-PAGE, and the gel was stained with Coomassie Blue.
Chromatin immunoprecipitation and DNA sequencing
Chromatin immunoprecipitation (ChIP) was performed in HeLa cells and HeLa cells expressing GFP-NKAP using a kit (ChIP-IT Express, 53008, Active Motif) according to the manufacturer’s protocol. Sequencing methods were based on Illumina protocols (Illumina Inc). Polyclonal GFP-specific antibodies and monoclonal NKAP antibodies (K85-80-5) were used for precipitation of NKAP. In controls, RNA Pol II and GFP-specific antibodies were used.
CLIP-seq and real-time polymerase chain reaction
FLAG-NKAP-transfected HEK-293 T cells were cultivated in 15-cm dishes in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum and antibiotics (streptomycin and penicillin). HEK 293T cells were irradiated with 300 mJ/cm2 ultraviolet (UV) light. The CLIP-seq was performed as described previously (27). Anti-FLAG antibodies were used for the immunoprecipitation. The immunoprecipitated and recovered RNA was used for the library preparation. The library preparation and sequencing was performed at the CCG (Cologne Center for Genomics). The CLIP raw reads were aligned to human genome version 19 using Bowtie. We further analyzed uniquely aligned reads using the University of California, Santa Cruz genome browser by running BEDtool utilities.
The total RNA was purified from control and knockdown cells. The purified RNA was used for real-time polymerase chain reaction (RT-PCR) to determine the levels of mRNA and pre-mRNA. Reverse transcription was carried out with an oligo(dT) primer and reverse transcriptase (Roche), according to the manufacturer’s instruction. The cDNA was then used for PCR reactions in the presence of pairs of forward and reverse primers (Supplementary Table S2). The relative amount for mRNA and pre-mRNA was quantified by Image J, which was then used to calculate the percentage of pre-mRNA.
RESULTS
NKAP shows punctuated distribution within the nucleus
NKAP is a 415 amino acid residues long highly basic protein of 47 kDa. The human and mouse proteins have an overall identity of 86%; the DUF 926 domains are identical. To have a better understanding of the localization and the role of the protein, we generated monoclonal antibodies against the DUF 926 domain of murine NKAP. mAb K85-80-5 recognized the mouse and the human protein in western blots and in immunofluorescence studies. In western blots, NKAP has an apparent molecular weight of ∼60 kDa, which might be the result of the unusually high content of charged residues (107 positively charged residues) or of posttranslational modifications such as phosphorylation. When we probed a blot containing protein lysates from several murine organs, we found that the protein is ubiquitously expressed and is particularly prominent in spleen, skin, testes, kidney and lung (Figure 1A). At the immunofluorescence level, endogenous NKAP was detected exclusively in the nucleus in a punctuated pattern in HeLa cells resembling nuclear speckles (Figure 1B).
Figure 1.
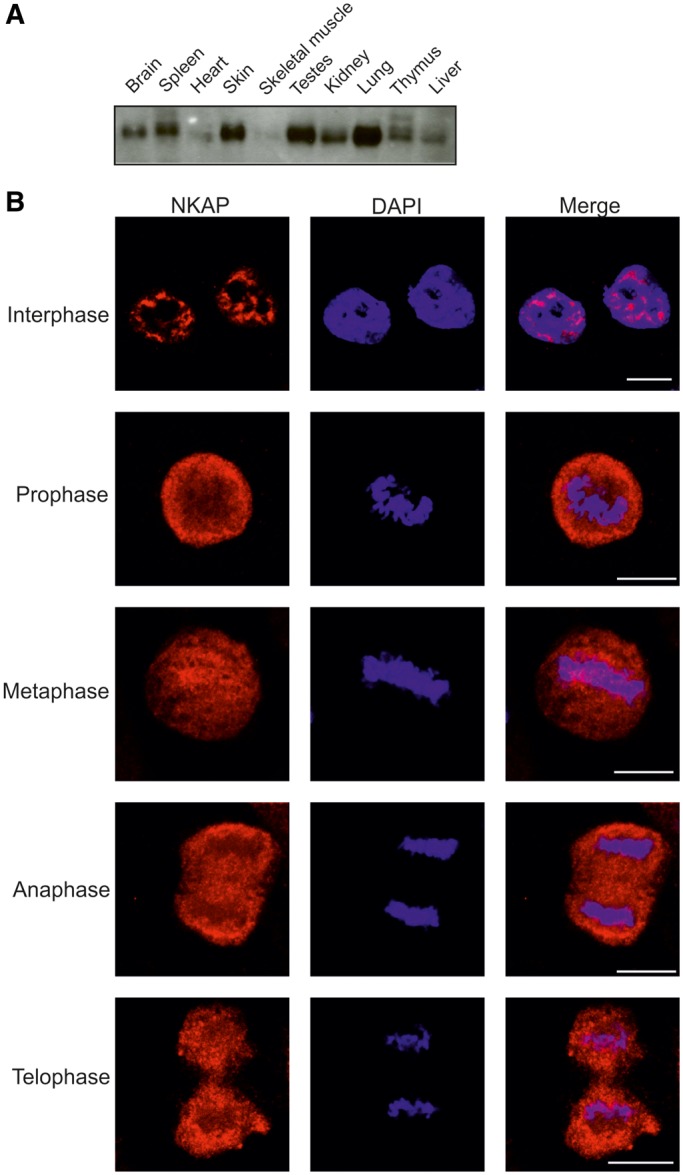
Expression and localization of NKAP. (A) Presence of NKAP in mouse tissues. Homogenates of adult mouse tissues as indicated were separated by SDS-PAGE (12% acrylamide) and the corresponding blot probed with NKAP-specific mAb K85-80-5. (B) Localization of NKAP during mitosis. HeLa cells were synchronized using nocodazole to block progression of the cell cycle and then released and fixed using 4% paraformaldehyde (PFA). Cells were stained with mAb K85-80-5, and nuclei (blue) were stained with 4′,6-diamidino-2-phenylindole (DAPI). The phases of the cell cycle are indicated. Bar, 10 µm.
On entry into mitosis and following the breakdown of the nuclear envelope, proteins associated with nuclear speckles become diffusely distributed throughout the cytoplasm. During metaphase, these proteins continue to localize in a diffuse cytoplasmic pattern. They then enter the daughter cells, and within 10 min they are present in the nucleus (28). To analyze the localization of NKAP during mitosis, we synchronized HeLa cells using nocodazole. After nocodazole release, we studied various phases of mitosis. We found that NKAP was diffusely distributed in the cytoplasm from prophase onward and remained cytoplasmic until telophase indicating that NKAP behaved like a speckle proteins (Figure 1B). Colocalization studies using SRSF1 antibodies showed that NKAP partially localized to mitotic interchromatin granule structures (Supplementary Figure S1).
The basic domain is required for the punctuate pattern of localization
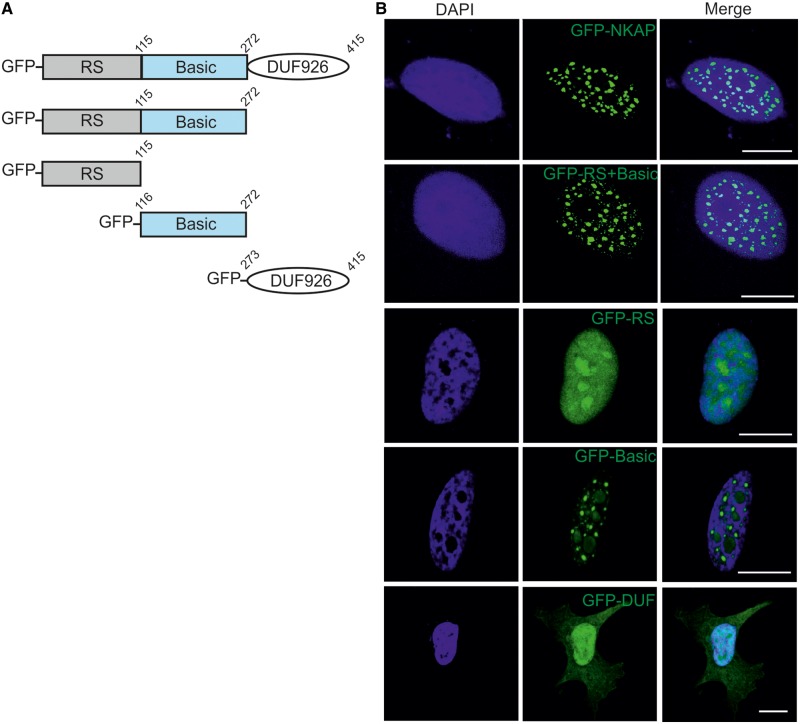
To narrow down the domain required for nuclear localization, we generated GFP-tagged full-length protein and deletion constructs GFP-RS+Basic, GFP-RS, GFP-Basic and GFP-DUF and expressed the proteins in HeLa cells (Figure 2A). For the full-length protein, we observed a nuclear-speckled pattern paralleling the distribution of the endogenous protein. GFP-RS+Basic encompassing the N-terminal domain of NKAP showed a similar localization (Figure 2B). This domain resembled the RS domains in SR proteins for which it is known that they are responsible for the speckle localization (11,12,13). However, the RS domain (GFP-RS) of NKAP did not show a nuclear-speckled pattern, demonstrating that, at least in this context, the RS domain is not responsible for punctuated pattern localization of NKAP. In contrast, the GFP-Basic domain showed punctuated localization like the full-length protein. The GFP-tagged DUF 926 domain was diffusely present in the nucleus and also showed weak cytosolic localization (Figure 2B).
Figure 2.
Subcellular localization of NKAP. (A) Schematic representation of NKAP polypeptides used in this study. (B) Full length NKAP, Rs + Basic domain, RS domain, Basic domain and DUF 926 domain were tagged with N-terminal GFP and expressed in HeLa cells that were fixed and stained with DAPI. Bar, 10 µm.
NKAP is a nuclear speckle protein
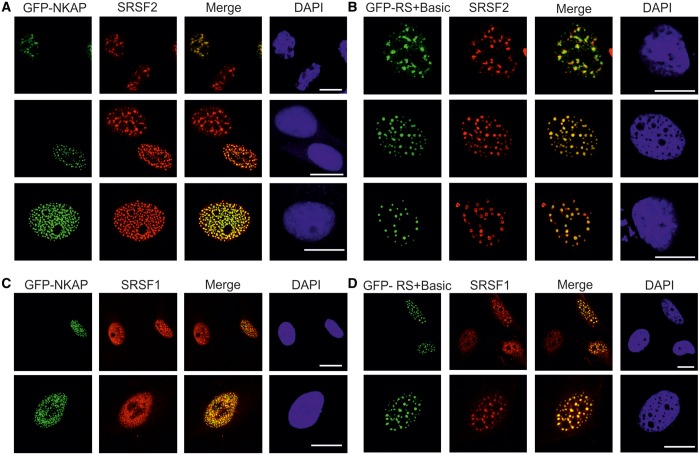
To address the subnuclear localization of NKAP, we performed immunofluorescence analysis for various intranuclear compartment markers using HeLa cells. Nuclear speckles were visualized by nuclear speckle marker SRSF2 and promyelocytic leukemia (PML) bodies by PML marker. NKAP co-localized with SRSF2, but not with PML bodies, suggesting its localization at nuclear speckles (Figure 3A). GFP-RS+Basic and GFP-Basic showed precise co-localization with SRSF2, whereas GFP-RS did not show co-localization with SRSF2. Thus, the RS domain of NKAP determines nuclear localization but not subnuclear speckle localization (Figure 3A).
Figure 3.
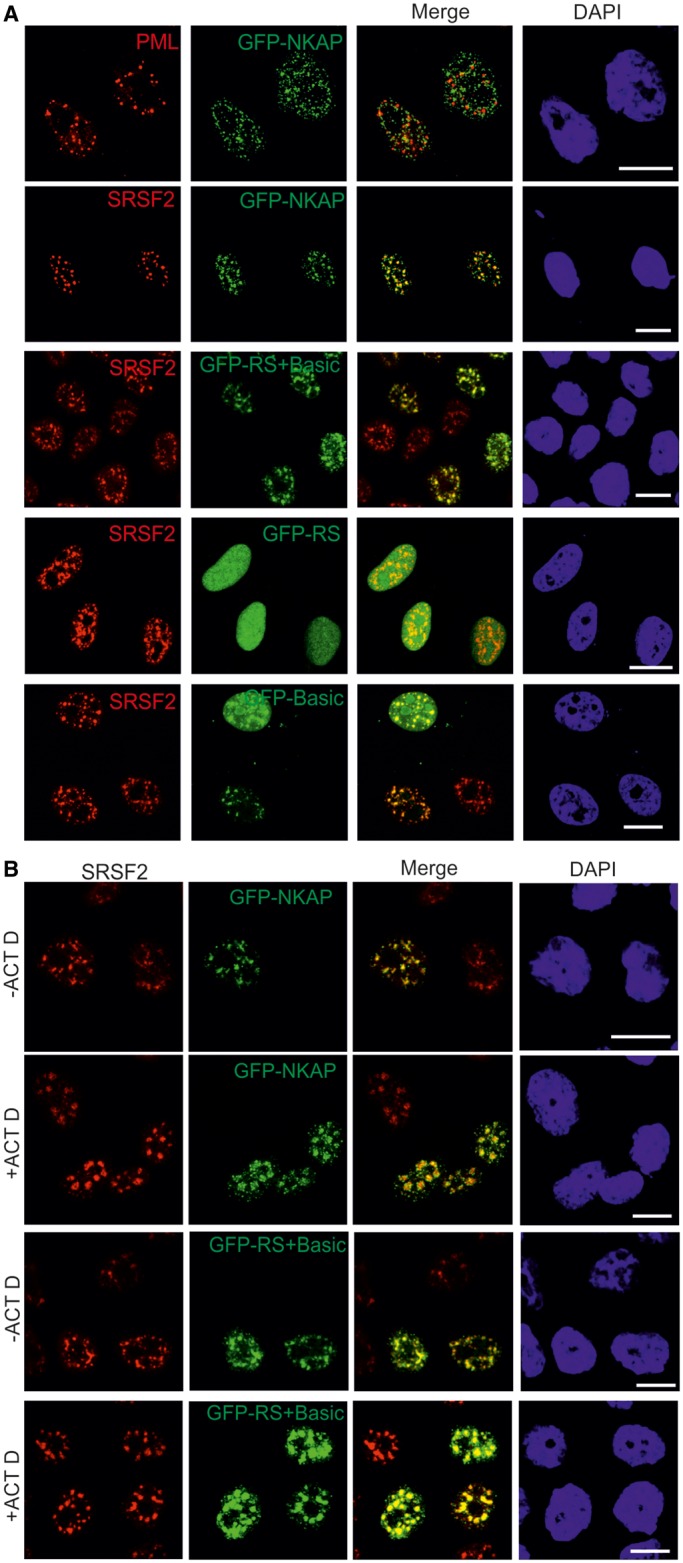
NKAP localizes to the nuclear speckles. (A) HeLa cells overexpressing GFP-NKAP, GFP-RS+Basic, GFP-RS and GFP-Basic were fixed with 4% PFA and stained for PML bodies and nuclear speckles by pAB PML and mAb SRSF2, respectively. DNA was stained with DAPI. (B) Arrest of transcription affects NKAP and RS + Basic distribution. HeLa cells expressing GFP-NKAP and GFP-RS+Basic were treated with actinomycin D, fixed with 4% PFA and stained for SRSF2. Bar, 10 µm.
Most of the nuclear speckle proteins accumulate in enlarged rounded spots when transcription is blocked (29). To check the possibility that NKAP shows a similar distribution on drug treatment, we studied the localization of NKAP after treatment with the transcription inhibitor actinomycin D. On inhibition of transcription, GFP-NKAP and GFP-RS+Basic showed rounded speckle localization, and the size of the speckles was increased (Figure 3B).
Ectopic expression of NKAP leads to altered nuclear speckle maintenance
Endogenous NKAP and GFP-NKAP showed a predominant localization in nuclear speckles, with little detectable diffuse nuclear signal (Figures 1B and 2). If NKAP is essential for nuclear speckle maintenance, we expected overexpression of NKAP to significantly alter the distribution of pre-mRNA processing factors. In fact, we observed that ectopic expression of NKAP in HeLa cells led to dramatically altered nuclear speckles organization as visualized by SRSF2 staining (Figure 4A). Usually 25–50 nuclear speckles were observed per nucleus during interphase in mammalian cells (1). The overexpression of NKAP led to an increase in nuclear speckle number (Figure 4A, bottom). On GFP-RS+Basic overexpression, we noted an alteration for SRSF2, which exhibited rounded speckles and in some cells, doughnut-shaped pattern surrounding GFP-RS+Basic (Figure 4B). It should be noted that a doughnut-shaped pattern has not been reported for SRSF2 overexpression (30). A similar NKAP overexpression effect was observed in case of SRSF1 localization (Figure 4C and D).
Figure 4.
Overexpression of NKAP and RS+Basic alters SRSF2 localization. (A) HeLa cells expressing GFP-NKAP were stained for SRSF2. Cells expressing moderate amounts (upper panel) and strongly overexpressing GFP-NKAP are shown (middle and lower panel). (B) Expression of GFP-RS+Basic in HeLa cells. Upper panel, cells expressing moderate amounts, middle and lower panel, overexpression of GFP-RS+Basic. Bar, 10 µm. (C) The distribution of SRSF1 and speckle number was altered when GFP-NKAP was strongly overexpressed. (D) Overexpression of RS+Basic led to altered localization of SRSF1.
NKAP is required for T-cell development and functions as transcriptional repressor for Notch signaling genes. Also, NKAP-deficient mice show a defect in T-cell maturation and the acquisition of functional competence. However, this effect of NKAP on T-cell maturation is independent of Notch (22). To test the direct involvement of NKAP in gene regulation, we performed a ChIP experiment. The ChIP was carried out for endogenous NKAP using mAb K85-80-5 antibody and also for exogenously expressed NKAP using GFP antibodies. For this, HeLa cells and HeLa cells overexpressing GFP-NKAP and GFP were fixed using formaldehyde and processed for the isolation and preparation of chromatin. Immunoprecipitation was performed using magnetic beads to which NKAP antibody, GFP antibodies and RNA PolII antibodies (positive control) were bound. GFP antibodies were used as negative control with which immunoprecipitation was carried out using a lysate from HeLa cells expressing GFP. Chromatin fragments were eluted from magnetic beads by reverse crosslinking. The precipitated DNA was purified and the presence and absence of chromatin determined by PCR amplification. Chromatin was successfully precipitated with endogenous NKAP and GFP-NKAP, whereas the negative control did not associate with chromatin (Supplementary Figure S2A and B). The precipitated DNA with GFP-NKAP was subsequently processed for sequencing. The ChIP-Seq data were analyzed and the sequences mapped onto the human genome (GRCH37/hg19) as described previously (31). The majority of the sequences that could be uniquely mapped represented centromere and other heterochromatic sequences (data not shown). In the previous study on NKAP function as transcriptional repressor, it was found associated with a Notch-regulated promoter through PCR analysis (20).
Identification and characterization of binding partners of NKAP
To understand the role of NKAP, we performed co-immunoprecipitation studies with HeLa cells expressing GFP-NKAP using GFP polyclonal antibodies. In this experiment, we were able to co-precipitate several proteins ranging from 30 to 130 kDa. The protein bands were cut out from gels and processed for LC-MS analysis. In this analysis, the majority of the proteins identified were RNA binding proteins, i.e. hnRNPs, RNA helicases and splicing factors (Supplementary Table S1). In further analysis, we wanted to confirm the mass spectrometry results and started to study the interaction of NKAP with the mRNA binding protein FUS/TLS (fused in sarcoma/translocated in sarcoma).
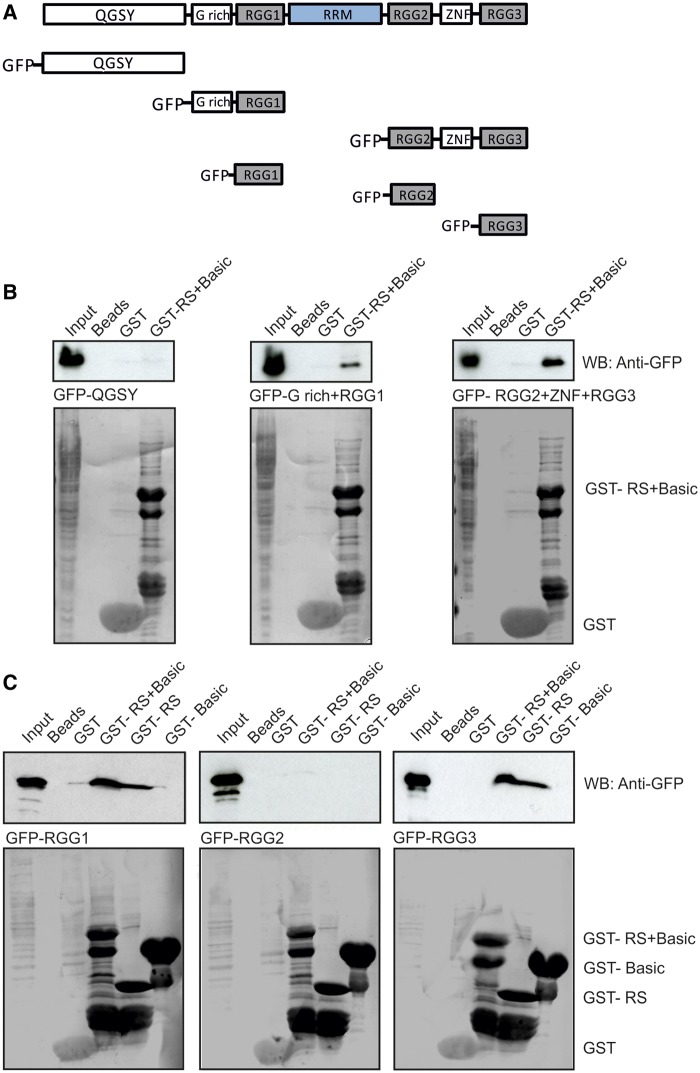
FUS/TLS is a member of the TET protein family. It contains an N-terminal QGSY region, a highly conserved RRM, multiple arginine/glycine-rich (RGG) repeats and a C-terminal zinc finger motif (Figure 6A). FUS/TLS was identified as hnRNP P2, an hnRNP involved in the maturation of pre-mRNA (32).
Figure 6.
The RS domain of NKAP interacts with RGG1 and RGG3. (A) Schematic representation of FUS/TLS constructs used in this study. (B) GST-RS+Basic pulls down GFP-G rich+RGG1 and GFP-RGG2+ZNF+RGG3 recognized by mAb GFP K184-3. GST-RS+Basic and GST were visualized by Ponceau S staining. Proteins were separated by 12% SDS-PAGE. (C) GST-RS domain of NKAP interacts with GFP-RGG1 and GFP-RGG3. GST-RS+Basic, GST-RS, GST-Basic and GST were visualized by Ponceau S staining.
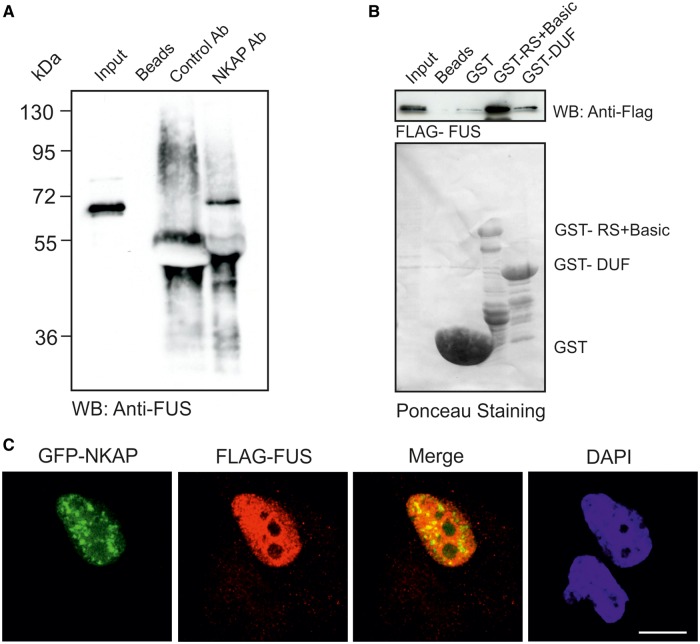
We analyzed the in vivo interaction of NKAP with FUS/TLS through co-immunoprecipitation using monoclonal NKAP antibodies. mAb K85-80-5 could co-precipitate endogenous FUS/TLS, which was detected by FUS antibody, confirming the interaction of endogenous NKAP and FUS/TLS (Figure 5A). We next narrowed down the association between NKAP and FUS in in vitro assays. We used GST-fusion proteins containing the RS+Basic of NKAP and the DUF 926 domain and carried out GST pull-down assays. Murine FUS cDNA was cloned and the protein expressed as FLAG fusion protein in HeLa cells. GST-RS+Basic could efficiently precipitate FLAG-tagged FUS, while weak signals were obtained for GST-DUF and negligible binding with the GST control (Figure 5B). We conclude that FUS and NKAP can interact either directly or indirectly and that the N-terminal domain of NKAP, which contains the RS domain, mediates the interaction. RS domains in general are known to control the interaction with other proteins and are important for protein–protein or protein–RNA interactions. Furthermore, we narrowed down the association site in FUS using different FUS polypeptides fused to GFP and expressed in HeLa cells (Figure 6A). GST-RS+Basic was used for pull downs. We found that the G-rich + RGG1 and the RGG2+ ZNF+ RGG3 domains of FUS could interact with RS+Basic, whereas GFP-QGSY did not, which indicated that FUS interacted with NKAP through RGG repeats (Figure 6B). To define the binding site in NKAP, we performed GST pull-down using different NKAP polypeptides fused to GST and GFP-fused RGG domains of FUS. The RS+Basic and RS domain of NKAP could pull down RGG1 and RGG3 but not RGG2, and GST-Basic showed no binding activity (Figure 6C).
Figure 5.
RS+Basic interacts with FUS. (A) Immunoprecipitation of FUS using anti-NKAP monoclonal antibody K85-80-5. (B) The GST-RS+Basic of NKAP precipitates FLAG-FUS recognized by polyclonal FLAG antibodies. GST, GST-RS+Basic and GST-DUF were visualized by Ponceau S staining. Proteins were separated by 12% SDS-PAGE. (C) HeLa cells were transfected with GFP-NKAP and FLAG-FUS, fixed and stained with FLAG antibodies. Secondary antibody conjugated with Alexa 568 was used. Nuclei were visualized with DAPI. Bar, 10 µm.
To analyze and compare the subcellular localization of FUS with NKAP, we performed co-immunofluorescence studies. Both proteins were located in the nucleus. FLAG-FUS exhibited a diffused pattern in addition to a speckled pattern. However, the FLAG-FUS staining was much more prominent than GFP-NKAP, and therefore clear co-localization was only seen in some areas (Figure 5C). Overexpression of NKAP and RS+Basic domain did not alter the localization of FUS/TLS (Supplementary Figure S3).
NKAP interacts with RNA
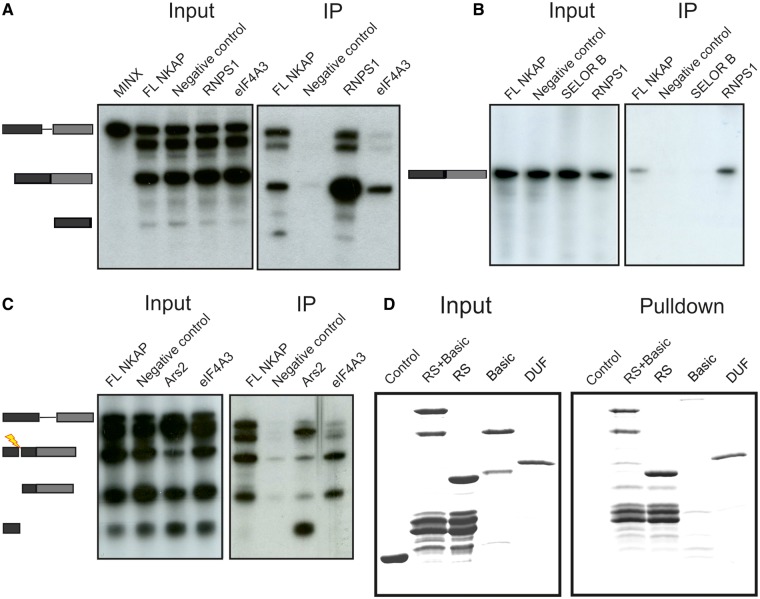
The presence of RNA binding proteins in co-immunoprecipitates of NKAP, the specific nuclear speckle localization and the presence of an RS domain at the N-terminus of NKAP prompted us to investigate the ability of NKAP to associate with RNA. To this end, we performed in vitro splicing experiments using NKAP as FLAG fusion protein expressed in HEK293T cells. The empty FLAG vector was used as a negative control and FLAG-tagged RNPS1 and eIF4A3 as positive controls. RNPS1 is a splicing activator and also a component of the exon junction complex (EJC) and interacts with pre-mRNA as well as spliced mRNA (33,34). eIF4A3 is a component of the EJC and preferentially associates with spliced mRNA (35). For the splicing assays, we used synthetic mRNA derived from adenovirus major late transcription unit (MINX), which is a single intron pre-mRNA (26). 32P-labeled MINX was spliced in vitro in different cell extracts, and immunoprecipitation was carried out using beads-coupled FLAG antibodies. Inputs were RNA from splicing reactions before immunoprecipitation. Both the input and immunoprecipitated RNA were resolved in denaturing 10% polyacrylamide gels. The results showed that NKAP interacted with pre-mRNA as well as with spliced mRNA (Figure 7A), indicating the ability of NKAP to interact with RNA. As expected, the positive control RNPS1 interacted with spliced mRNA and pre-mRNA, whereas eIF4A3 only precipitated spliced mRNA (Figure 7A).
Figure 7.
NKAP interacts with RNA. (A) 32P-labeled MINX pre-mRNA was spliced in vitro in HEK293T cell extracts containing FLAG-tagged proteins (RNPS1 and eIF4A3 as positive control, FLAG alone served as negative control) followed by immunoprecipitation by FLAG antibody coupled beads (IP). Aliquots of the total reactions were also analyzed (input). The positions of pre-mRNA, spliced mRNA and exon 1 (from top to bottom) are shown on the left. (B) 32P-labeled intronless mRNA MINX was incubated for 2 h in a HEK293T cell extract expressing FLAG-tagged proteins (RNPS1 as positive control, SELOR B and FLAG were negative control) followed by immunoprecipitation using FLAG antibodies coupled beads (IP). Aliquots of the total reactions were also analyzed (input). (C) 32P-labeled MINX pre-mRNA was spliced in vitro using HEK293T cell extracts expressing FLAG-tagged proteins. It was followed by addition of an oligonucleotide complementary to the first exon to activate RNase H cleavage of the RNA and to release a 5′ cap containing fragment and by immunoprecipitation with FLAG antibodies (IP). Aliquots of the total reactions were also analyzed using denaturing PAGE (Input). The mobilities of pre-mRNA, spliced mRNA, exon 1 and 5′ fragment are marked on the left. (D) RNA-binding assay using Sepharose-conjugated RNA homopolymers and GST-RS+Basic, GST-RS and GST-DUF followed by western blotting. GST was used for control.
Further, we next used the intronless MINX to test whether NKAP binds splicing independently to mRNA. We used FLAG-tagged RNPS1 because it binds intronless RNA. FLAG-tagged SELOR B (speckle localizer and RNA binding domain of Barentsz) domain was used as negative control, as it binds mRNA but not intronless mRNA. 32P-labeled intronless MINX was added to the various splicing reactions. After the incubation, FLAG-tagged proteins were precipitated with FLAG antibody coupled beads and the co-precipitated RNA was analyzed by denaturing PAGE. NKAP bound intronless mRNA, although the binding was weak as compared with the positive control RNPS1 (Figure 7B).
Although NKAP interacts with pre-mRNA and intronless mRNA, the specific function of NKAP in RNA processing is unclear. Therefore, we next aimed to identify the possible binding site of NKAP on the RNA. In vitro splicing was performed by adding 32P-labeled MINX to different cell lysates followed by addition of a complementary DNA-oligo to the 5′-end of the exon, which cleaves DNA/RNA duplexes to release the 5′-end of the RNA including the cap. An immunoprecipitation was performed, and co-immunoprecipitated RNA was analyzed by denaturing PAGE. The positive control, mRNA 5′-cap binding protein Ars2, interacted with pre-mRNA and with the 5′ fragment of the RNA. In contrast, NKAP interacted with all RNAs, indicating that NKAP does not preferentially interact with the 5′-cap structure or a particular region of the mRNA (Figure 7C).
We next investigated a direct RNA binding ability of NKAP using two synthetic RNA homopolymers, poly(C) and poly(U), coupled to Sepharose beads. This method has been previously proven to be suitable for studying RNA binding properties of RNA binding proteins (36). The GST-fused RS+Basic, RS and DUF polypeptides displayed binding to poly(U)-Sepharose beads (Figure7D), but no binding to poly(C) beads (data not shown), which indicates that it does not belong to the group of RNA binding proteins that have a high affinity for poly(C). GST-Basic did not show any binding to poly(U) or poly(C) beads and neither did GST, indicating a specific affinity of NKAP for RNA.
Genome-wide analysis of NKAP–RNA interaction using CLIP-seq
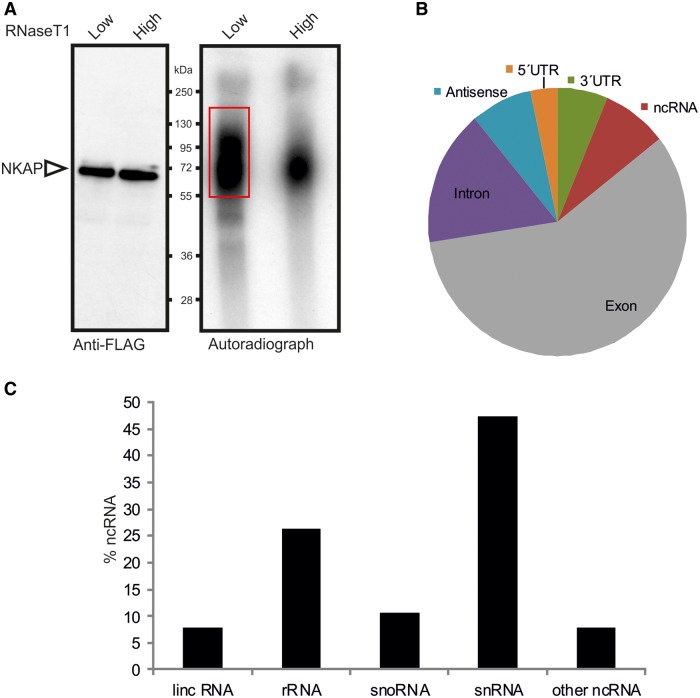
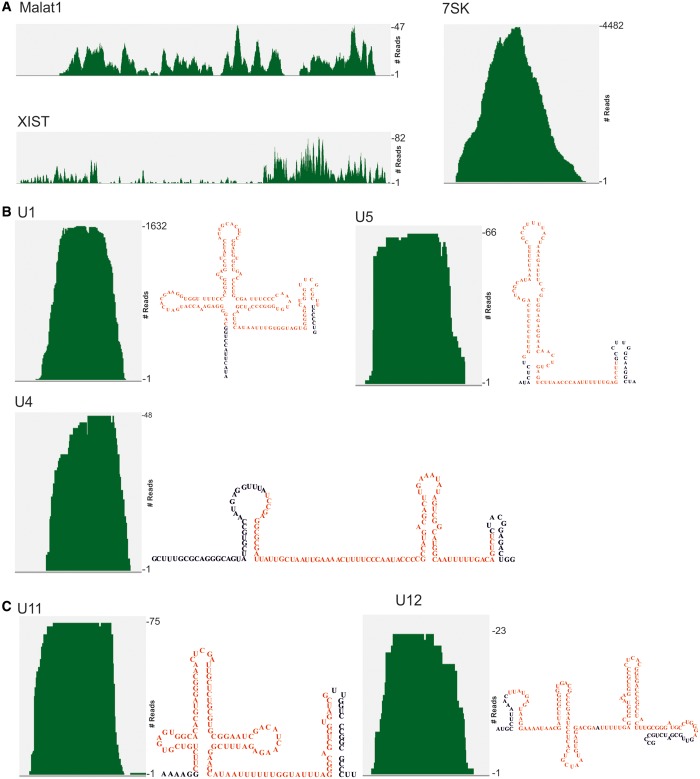
In human embryonic kidney cells, a photoreactive nucleoside-enhanced UV crosslinking and oligo (dT) affinity purification approach had been used previously to identify the mRNA-bound proteome, and NKAP was listed as one of the novel RNA binding protein (37). We used the CLIP method to identify the in vivo RNA-binding profile in HEK 293T cells. For this purpose, we UV-irradiated FLAG-NKAP-transfected HEK 293T cells to induce crosslinking between proteins and nucleic acids, followed by immunoprecipitation using anti-FLAG antibodies, while control non-UV-treated cells produced no signal (data not shown). We isolated NKAP–RNA complex from low RNase T1-treated experiment (Figure 8A). Crosslinked immunopurified RNA was digested to lengths of 70–100 nt, reverse transcribed and prepared for next-generation sequencing. The resulting reads were mapped to the human genome. In total, CLIP produced 257 451 unique reads for NKAP. The mapped tags were distributed among diverse primary RNA transcripts with the majority mapping to exonic regions of pre-mRNA (Figure 8B). We next analyzed for specific NKAP-binding motifs with MEME software (http://nbcr-222.ucsd.edu/opal2/services/MEME_4.9.1), but detected variable motifs that widely varied by adding or eliminating different parameters. Interestingly, we also detected NKAP binding to various ncRNAs. The most abundant ncRNA classes in CLIP tags were snRNAs and rRNAs (Figure 8B). Similar to other SR proteins, NKAP crosslinked to the 7SK RNA known to play a critical role as a molecular sink for transcription elongation regulators (38), with Malat-1 that co-localizes to nuclear speckles (39), XIST, which is an X-inactive-specific transcript (Figure 9A) and with numerous other ncRNAs. These findings raise the possibility that the previously recorded functions of NKAP in transcription regulation may be mediated through its interaction with various regulatory ncRNAs.
Figure 8.
Global analysis of NKAP binding to RNA by CLIP-seq. (A) Autoradiograph of cross-linked NKAP-RNA complexes using denaturing gel electrophoresis and membrane transfer. RNA was partially digested using low or high concentration of RNase T1. The marked area was cut from the membrane and subjected to protocols for the isolation of RNA, which was further used for RNA-seq. (B) Distributions of NKAP CLIP-tags. Binding regions are mapped to exons, UTRs, introns, antisense and ncRNAs according to the University of California, Santa Cruz human genome browser. Pie-charts show ratios of binding regions mapped to the indicated regions. (C) Distribution of cross-link sites within the ncRNA classes.
Figure 9.
Association of NKAP with ncRNA. (A) NKAP association with Malat1, Xist and 7SK. (B and C) Interaction of NKAP with snRNAs required for splicing of both major (B) and minor (C) classes of introns. The secondary structure of each snRNA is shown on the right. The covered regions in CLIP-seq experiment of snRNAs are shown in red.
Specifically pertinent to our current investigation of NKAP in RNA biogenesis, we found binding with many snRNAs required for splicing (Figure 9B). As U1 snRNA is the most abundant one in the cell, the tag density was higher for U1 snRNA. The tag density on U4 and U5 snRNA was relatively equal. In addition, NKAP seemed to prefer U11 over U12 and because of this it may be involved in splicing of minor introns (Figure 9C). Notably, snRNAs for minor introns are lower in abundance compared with major introns, indicating that NKAP plays a possible role in minor intron splicing, which will be addressed in the future. The snRNA and NKAP interaction might play an important role in regulation of snRNP biogenesis/maturation and/or assembly into the spliceosome.
The role of NKAP in splicing
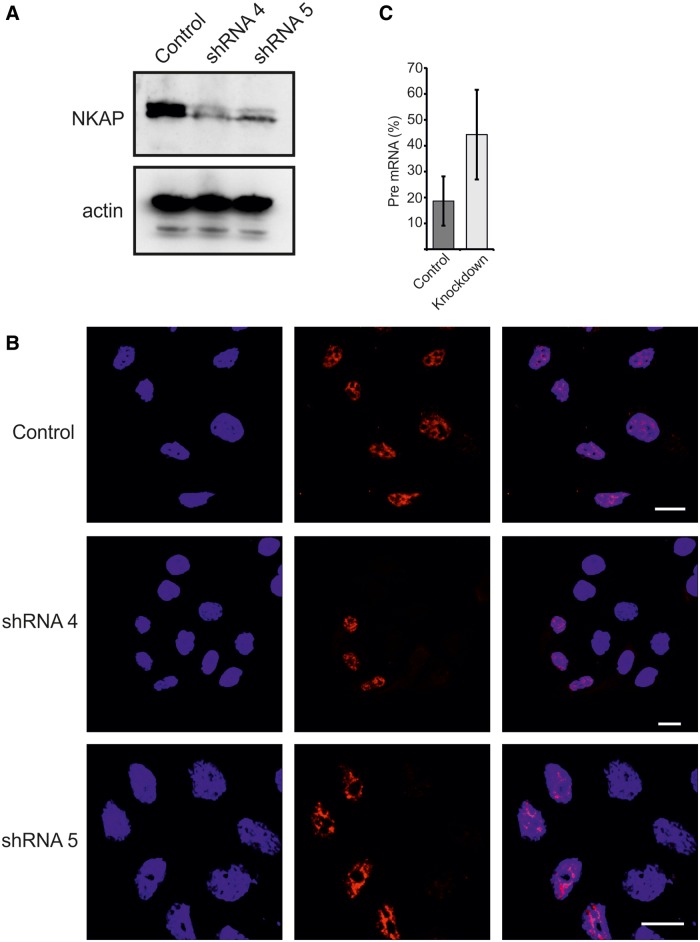
To analyze whether NKAP is required for splicing, we used two different small hairpin RNAs to knockdown NKAP. We confirmed the efficiency of knockdown by western blot and immunofluorescence (Figure 10A and B), which clearly showed reduced levels of NKAP when compared with the control. In a large-scale proteomic analysis of spliceosomal complexes, NKAP was found with different spliceosomal complexes, but the role of NKAP in these complexes and in splicing is not understood (40–43).To address this, we performed RT-PCR to quantify the pre-mRNA and mRNA ratio of the house-keeping gene glyceraldehyde 3-phosphate dehydrogenase (GAPDH). We always had variable knockdown efficiency, and therefore in some experiments, we did not observe a large difference in control and knockdown pre-mRNA percentage. This leads to big error bars in the control and knockdown pre-mRNA percentage. Nevertheless, the percentage of GAPDH pre-mRNA [pre-mRNA/(mRNA + pre-mRNA)] was significantly increased (n = 6, P = 0.0164) (Figure 10C). Taken together, our data demonstrate that ablation of NKAP strongly affects pre-mRNA splicing.
Figure 10.
Role of NKAP in regulating splicing. (A) Efficient knockdown of NKAP using two different oligos. Western blotting was performed using NKAP mAB K85-80-5. (B) Control and knockdown HEK 293T cells were fixed with 4% PFA and stained using mAB K85-80-5. (C) The pre-mRNA percentage of GAPDH in control and NKAP knockdown HEK 293T cells. A significant increase was observed in knockdown cells (n = 6, P = 0.0164).
DISCUSSION
Many pre-mRNA splicing factors including snRNPs and SR proteins were found at nuclear speckles (44). SR proteins are highly conserved and characterized by serine arginine repeats, the RS domain, which is a signature of nuclear speckle proteins. NKAP carries this signature at the N-terminus suggesting that NKAP is an SR-related protein. The RS domain has been shown to be important for the speckle localization. However, the RS domain of NKAP is neither necessary nor sufficient for speckle localization. Although the RS domain has a nuclear localization signal, targeting to the nuclear speckle requires the basic domain of NKAP that contains lysine repeats. Other regions in specific proteins have also been reported to act as speckle-localizing sequences like the threonine–proline repeats in SF3B1/SF3b155 (45) and the ‘Forkhead Associated’ domain in PPP1R8/NIPP1 (46). Furthermore, histidine repeats have been described as a targeting signal for the nuclear speckle (47). However, we did not find a specific sequence in the basic domain mediating the targeting to nuclear speckles.
On inhibition of RNAP II transcription, speckles become round and increase in size, whereas diffuse speckles disappear. These changes are caused presumably by a splicing arrest resulting from transcriptional inhibition. All splicing factors then leave the nucleoplasm and accumulate in the speckles (30). Cells treated with actinomycin D showed redistribution of both SRSF2 and NKAP to enlarged and rounded speckles and a complete loss of diffuse nuclear speckle staining. A similar effect was seen in case of GFP-RS+Basic. On entry into mitosis, all pre-mRNA processing factors diffusely distributed throughout the cytosol and reassemble into speckles in telophase. NKAP also showed a similar pattern of localization during mitosis. These results support that NKAP behavior is reminiscent with other SR proteins.
The overexpression of NKAP leads to an increase in speckle number. The increase in speckle number differs from what has been found in previous studies. Normally, overexpression of SR proteins, i.e. RNPS1, causes enlarged nuclear speckle structures but not a dramatic increase in speckle number but rather a reduction (48). NKAP may not be important for the nuclear speckle organization, as it did not co-localize with the mitotic interchromatin granule marker SRSF1 during mitosis. Our results suggest that NKAP has an important role in maintenance of nuclear speckles. Overexpression of GFP-RS+Basic did not lead to a dramatic increase in speckle number and also showed doughnut-like structures where SRSF2 and SON surrounded GFP-RS+Basic (Supplementary Figure S4). Similar structures were observed on depletion of SON, a protein involved in controlling cell-cycle progression through SON-dependent constitutive splicing at suboptimal splice sites (49). Normally, EJC components are enriched in the doughnut-shaped regions, also called perispeckles, which surround nuclear speckles. Perispeckles are major locations for mRNA transcription, splicing and EJC assembly. Splicing factor SRSF2 has not been detected so far in the perispeckle region (50). One proposal is that RS+Basic promotes the localization of splicing factors to these perispeckles and that the DUF 926 domain might be controlling the function of NKAP. The in vitro splicing assay for NKAP demonstrated that NKAP interacted with RNA and bound to pre-mRNA and spliced mRNA suggesting that the protein could modulate splicing in vivo by recruiting, activating or stabilizing RNA processing factors. Using synthetic homopolymers, we have found that the RS domain and DUF 926 domain of NKAP act as RNA binding domain. The RS domain in general has been shown to interact with pre-mRNA; specifically it contacts a 10-nt region surrounding the branch point (17). In addition to this, the interaction of DUF 926 domain with RNA is an intriguing finding. We hypothesize that the DUF 926 interaction with RNA might enhance the RNA and RS domain interaction or the DUF 926 domain might be a controlling factor for the RNA–RS domain interaction.
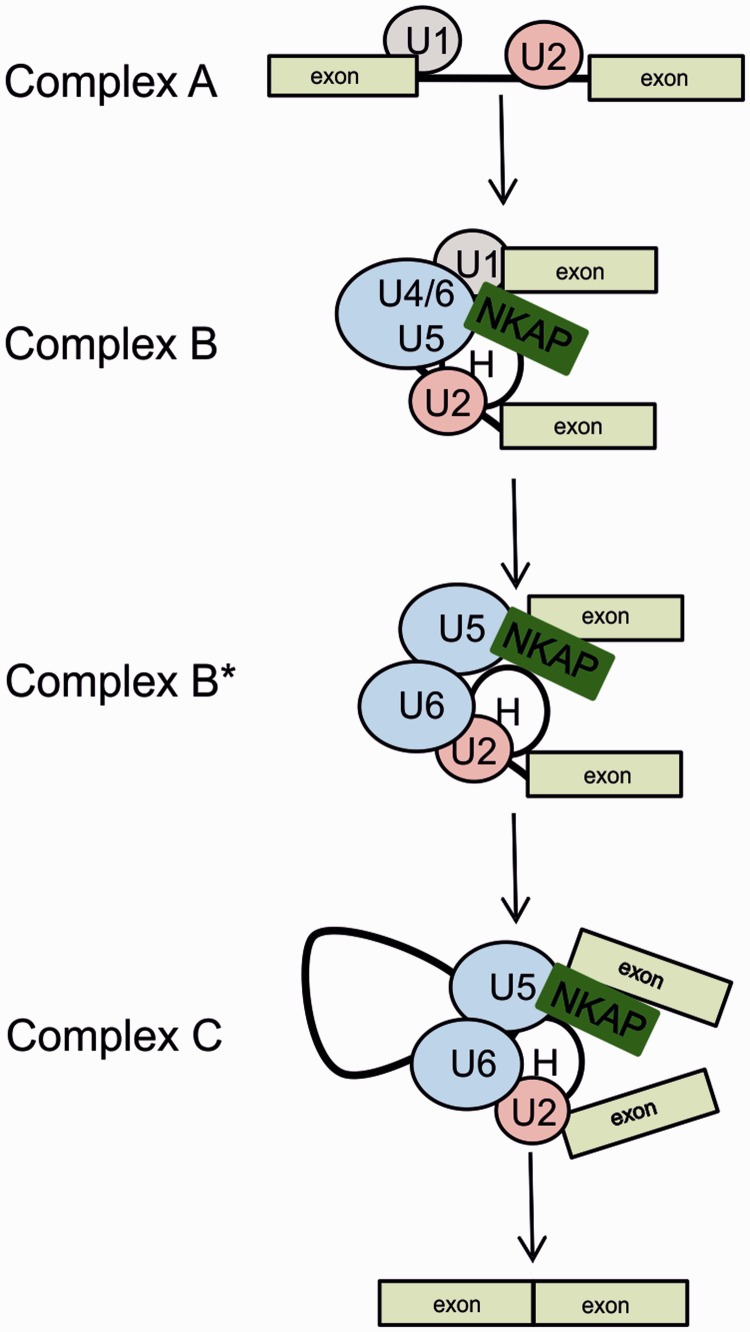
In Drosophila, a large-scale yeast two-hybrid screen was performed so that a proteome-wide protein interaction map could be defined, and three potential binding partners for CG6066 (the Drosophila NKAP ortholog) were found. These previously uncharacterized proteins (CG14323, CG6843 and CG31211) were linked to splicing components through an extensive set of interactions. Although these proteins have no recognizable RNA binding motif, the degree of high-confidence connectivity with other splicing components suggested that they were splicing complex members (23). There were further hints for a link with the spliceosomal complex. For instance, NKAP was found associated with specific human spliceosomal complexes such as B* complex, C-complex and P complex (40–43). The CLIP analysis revealed the global landscape of NKAP–RNA interactions including its direct binding on snRNAs. Mapping of U1 and U4 snRNA in NKAP CLIP-Seq is more intriguing, as they contribute to the formation of complex B during splicing. More details of this interaction need to be addressed, specifically, NKAP recruitment to the B complex and its possible trigger to release U1 and U4 snRNAs leading to the formation of the B* complex (Figure 11).
Figure 11.
Assembly of spliceosomal complexes and proposed function of NKAP. Model presenting the function of NKAP and its association with snRNAs. NKAP associates with the B splicing complex, specifically with U1, U4/U5 snRNA and complex H (hnRNP proteins) and could act as a trigger to activate this complex via dissociation of U1 and U4 snRNA while keeping its tight association with U5 snRNA and complex H.
Additionally, in our co-immunoprecipitation studies, many RNA binding proteins have been detected as potential interacting partners for NKAP such as FUS/TLS, hnRNPs and RNA helicases for which we could confirm the interaction by independent methods.
Proteomic analysis identified FUS previously as part of the spliceosome machinery (51,52). FUS/TLS associates in vitro with large transcription-splicing complexes that bind the 5′ splice sites of pre-mRNA (53) and has been also proposed to directly bind the pre-mRNA 3′ splice site (54). Furthermore, FUS/TLS associates with other splicing factors including YB-1, SR proteins SRSF2 and TASR, polypyrimidine tract-binding protein or hnRNP I and C1/C2 (55–59).
The N terminus of FUS contains a glutamine-, serine- and tyrosine-rich region that functions as a transcriptional activation domain when fused to a heterologous DNA binding domain (60). This is followed by multiple domains involved in interactions with RNA: an RRM motif flanked by several RGG boxes and a zinc finger. NKAP is divided into approximately three domains, N-terminal RS repeats, a highly basic middle domain and the C-terminal DUF 926. The RS domain in general has been shown to interact with several proteins (16). We have shown that the RS domain of NKAP mediates protein–protein interaction with RGG1 and RGG3 repeats of FUS in GST pull-down assay (Figure 8). RGG boxes in hnRNP U, Ewing sarcoma breakpoint region 1 (EWSR1, also EWS), FUS and fragile X mental retardation 1 (FMR1, also FMRP) were reported to mediate interactions with RNA (60–64). RGG-boxes are also able to mediate protein–protein interactions (65). For instance, the RGG boxes in FUS interact with SR proteins (66). RGG repeats have important roles in polyribosome association where the different arginines of the RGG box are important for the binding of different RNAs (67). Importantly, RGG repeats contribute toward enhancing the RRM binding affinity for RNA (68). FUS is a complex H protein and has also been found in E, A, B and C splicing complexes (40,43,69–72). It remains possible that the association of NKAP with the RGG repeats facilitates the binding of FUS with RNA during splicing. FUS is also a regulator of alternative splicing events in a position-dependent manner (73). The NKAP–FUS interaction might well be significant for such alternative splicing events, which should be addressed further.
Like FUS/TLS, NKAP is expressed in almost all organs. RT-PCR and immunofluorescence studies revealed that NKAP is highly expressed in mouse brain from the embryo to adult stage (data not shown). Mutations in FUS/TLS cause an inherited form of the neurodegenerative disease amyotrophic lateral sclerosis (ALS) (74). ALS is an adult-onset neurodegenerative disorder in which premature loss of motor neurons leads to fatal paralysis. Whether NKAP and FUS/TLS operate in common biochemical signaling pathways in ALS is not known. The role of FUS in RNA processing has been shown in association with ALS and other neurodegenerative diseases. Here we hypothesize that the NKAP and FUS interaction may regulate RNA processing together, and this might be a crucial factor in ALS, which must be addressed further.
Based on our data, we present a model in which NKAP associates with B complex, specifically to U1 and U4/U5 snRNA, and could act as a trigger to activate this complex via dissociation of U1 and U4 snRNA while keeping its tight association with U5 snRNA. Its interaction with FUS, a component of complex H, may have a critical role in this activation event. In complex C, where pre-mRNAs are spliced, NKAP still associates with U5 snRNA and FUS, which leads to the final spliced transcript. In addition to this, the NKAP–FUS interaction could also facilitate alternative splicing.
Taken together, our current study reveals NKAP as novel RNA binding protein with structural features similar to SR-related proteins. We further delineate its function with regard to pre-mRNA processing based on its subnuclear speckled distribution, physical interaction with RNA binding proteins and in vitro RNA splicing assays. CLIP-seq revealed that NKAP binds to many different classes of ncRNA, consistent with its tight association with complex H proteins. Importantly, our study revealed a key role for NKAP in regulating constitutive splicing. Therefore, our work has provided a novel function of NKAP in RNA splicing.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
The authors thank Dr Gerrit Praefcke, Institute of Genetics, University of Cologne, for providing PML antibodies. B.D.B. and A.S. were supported by the IGSDHD. This work was supported by CECAD. The funding through the Maria Pesch Foundation to B.D.B. is acknowledged. RNA seq was carried out at the Cologne Center for Genomics (CCG). The authors also thank Prof. Jernej Ule, UCL, London, for his suggestions in optimizing CLIP experiment. They also thank Prof. Xiang-Dong Fu, UCSD, USA, Prof. Don Cleveland, UCSD, USA, and Prof. Jie Huang, Wuhan University, China, for their help in analysis.
FUNDING
Cologne Excellence Cluster on Cellular Stress Responses in Aging-Associated Diseases (CECAD), International Graduate School in Development, Health and Disease (IGSDHD), and Maria Pesch Foundation. Funding for open access charge: Institute for Biochemistry I, Medical Faculty, University of Cologne.
Conflict of interest statement. None declared.
REFERENCES
- 1.Lamond AI, Spector DL. Nuclear speckles: a model for nuclear organelles. Nat. Rev. Mol. Cell Biol. 2003;4:605–612. doi: 10.1038/nrm1172. [DOI] [PubMed] [Google Scholar]
- 2.Spector DL, Lamond AI. Nuclear speckles. Cold Spring Harb. Perspect. Biol. 2011;3:a000646. doi: 10.1101/cshperspect.a000646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lafarga M, Casafont I, Bengoechea R, Tapia O, Berciano MT. Cajal's contribution to the knowledge of the neuronal cell nucleus. Chromosoma. 2009;118:437–443. doi: 10.1007/s00412-009-0212-x. [DOI] [PubMed] [Google Scholar]
- 4.Hall LL, Smith KP, Byron M, Lawrence JB. Molecular anatomy of a speckle. Anat. Rec. A Discov. Mol. Cell. Evol. Biol. 2006;288:664–675. doi: 10.1002/ar.a.20336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Melcák I, Cermanová S, Jirsová K, Koberna K, Malínský J, Raska I. Nuclear pre-mRNA compartmentalization: trafficking of released transcripts to splicing factor reservoirs. Mol. Biol. Cell. 2000;11:497–510. doi: 10.1091/mbc.11.2.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Melcák I, Melcáková S, Kopský V, Vecerová J, Raska I. Prespliceosomal assembly on microinjected precursor mRNA takes place in nuclear speckles. Mol. Biol. Cell. 2001;12:393–406. doi: 10.1091/mbc.12.2.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spector DL, Schrier WH, Busch H. Immunoelectron microscopic localization of snRNPs. Biol. Cell. 1983;49:1–10. doi: 10.1111/j.1768-322x.1984.tb00215.x. [DOI] [PubMed] [Google Scholar]
- 8.Spector DL, Fu XD, Maniatis T. Associations between distinct pre-mRNA splicing components and the cell nucleus. EMBO J. 1991;10:3467–3481. doi: 10.1002/j.1460-2075.1991.tb04911.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shopland LS, Johnson CV, Byron M, McNeil J, Lawrence JB. Clustering of multiple specific genes and gene-rich R-bands around SC-35 domains: evidence for local euchromatic neighborhoods. J. Cell Biol. 2003;162:981–990. doi: 10.1083/jcb.200303131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Raska I. Nuclear ultrastructures associated with the RNA synthesis and processing. J. Cell. Biochem. 1995;59:11–26. doi: 10.1002/jcb.240590103. [DOI] [PubMed] [Google Scholar]
- 11.Cáceres JF, Misteli T, Screaton GR, Spector DL, Krainer AR. Role of the modular domains of SR proteins in subnuclear localization and alternative splicing specificity. J. Cell Biol. 1997;138:225–238. doi: 10.1083/jcb.138.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hedley ML, Amrein H, Maniatis T. An amino acid sequence motif sufficient for subnuclear localization of an arginine/serine-rich splicing factor. Proc. Natl Acad. Sci. USA. 1995;92:524–528. doi: 10.1073/pnas.92.25.11524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li H, Bingham PM. Arginine/serine-rich domains of the su(wa) and tra RNA processing regulators target proteins to a subnuclear compartment implicated in splicing. Cell. 1991;67:335–342. doi: 10.1016/0092-8674(91)90185-2. [DOI] [PubMed] [Google Scholar]
- 14.Graveley BR. Sorting out the complexity of SR protein functions. RNA. 2000;6:1197–1211. doi: 10.1017/s1355838200000960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Long JC, Caceres JF. The SR protein family of splicing factors: master regulators of gene expression. Biochem. J. 2009;417:15–27. doi: 10.1042/BJ20081501. [DOI] [PubMed] [Google Scholar]
- 16.Wu JY, Maniatis T. Specific interactions between proteins implicated in splice site selection and regulated alternative splicing. Cell. 1993;75:1061–1070. doi: 10.1016/0092-8674(93)90316-i. [DOI] [PubMed] [Google Scholar]
- 17.Shen H, Kan JL, Green MR. Arginine-serine-rich domains bound at splicing enhancers contact the branchpoint to promote prespliceosome assembly. Mol. Cell. 2004;13:367–376. doi: 10.1016/s1097-2765(04)00025-5. [DOI] [PubMed] [Google Scholar]
- 18.Lin S, Coutinho-Mansfield G, Wang D, Pandit S, Fu XD. The splicing factor SC35 has an active role in transcriptional elongation. Nat. Struct. Mol. Biol. 2008;15:819–826. doi: 10.1038/nsmb.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen D, Li Z, Yang Q, Zhang J, Zhai Z, Shu HB. Identification of a nuclear protein that promotes NF-kappaB activation. Biochem. Biophys. Res. Commun. 2003;310:720–724. doi: 10.1016/j.bbrc.2003.09.074. [DOI] [PubMed] [Google Scholar]
- 20.Pajerowski AG, Nguyen C, Aghajanian H, Shapiro MJ, Shapiro VS. NKAP is a transcriptional repressor of notch signaling and is required for T cell development. Immunity. 2009;30:696–707. doi: 10.1016/j.immuni.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pajerowski AG, Shapiro MJ, Gwin K, Sundsbak R, Nelson-Holte M, Medina K, Shapiro VS. Adult hematopoietic stem cells require NKAP for maintenance and survival. Blood. 2010;116:2684–2693. doi: 10.1182/blood-2010-02-268391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hsu FC, Pajerowski AG, Nelson-Holte M, Sundsbak R, Shapiro VS. NKAP is required for T cell maturation and acquisition of functional competency. J. Exp. Med. 2011;208:291–304. doi: 10.1084/jem.20101874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Giot L, Bader JS, Brouwer C, Chaudhuri A, Kuang B, Li Y, Hao YL, Ooi CE, Godwin B, Vitols E, et al. A protein interaction map of Drosophila melanogaster. Science. 2003;302:1727–1736. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- 24.Hegele A, Kamburov A, Grossmann A, Sourlis C, Wowro S, Weimann M, Will CL, Pena V, Lührmann R, Stelzl U. Dynamic protein-protein interaction wiring of the human spliceosome. Mol. Cell. 2012;45:567–580. doi: 10.1016/j.molcel.2011.12.034. [DOI] [PubMed] [Google Scholar]
- 25.Peche V, Shekar S, Leichter M, Korte H, Schroder R, Schleicher M, Holak TA, Clemen CS, Ramanath YB, Pfitzer G, et al. CAP2, cyclase-associated protein 2, is a dual compartment protein. Cell. Mol. Life Sci. 2007;64:2702–2715. doi: 10.1007/s00018-007-7316-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gehring NH, Lamprinaki S, Hentze MW, Kulozik AE. The hierarchy of exon-junction complex assembly by the spliceosome explains key features of mammalian nonsense-mediated mRNA decay. PLoS Biol. 2009;7:e1000120. doi: 10.1371/journal.pbio.1000120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Konig J, Zarnack K, Rot G, Curk T, Kayikci M, Zupan B, Turner DJ, Luscombe NM, Ule J. iCLIP-transcriptome-wide mapping of protein-RNA interactions with individual nucleotide resolution. J. Vis. Exp. 2011;50:e2638. doi: 10.3791/2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prasanth KV, Sacco-Bubulya P, Prasanth SG, Spector DL. Sequential entry of components of gene expression machinery into daughter nuclei. Mol. Biol. Cell. 2003;14:1043–1057. doi: 10.1091/mbc.E02-10-0669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O’Keefe RTA, Mayeda A, Sadowski CL, Krainer AR, Spector DL. Disruption of pre-mRNA splicing in vivo results in reorganization of splicing factors. J. Cell. Biol. 1994;124:249–260. doi: 10.1083/jcb.124.3.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tripathi K, Parnaik VK. Differential dynamics of splicing factor SC35 during the cell cycle. J. Biosci. 2008;33:345–354. doi: 10.1007/s12038-008-0054-3. [DOI] [PubMed] [Google Scholar]
- 31.Rashmi RN, Eckes B, Glöckner G, Groth M, Neumann S, Gloy J, Sellin L, Walz G, Schneider M, Karakesisoglou I, et al. The nuclear envelope protein Nesprin-2 has roles in cell proliferation and differentiation during wound healing. Nucleus. 2012;3:172–186. doi: 10.4161/nucl.19090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Calvio C, Neubauer G, Mann M, Lamond AI. Identification of hnRNP P2 as TLS/FUS using electrospray mass spectrometry. RNA. 1995;1:724–733. [PMC free article] [PubMed] [Google Scholar]
- 33.Mayeda A, Badolato J, Kobayashi R, Zhang MQ, Gardiner EM, Krainer AR. Purification and characterization of human RNPS1: a general activator of pre-mRNA splicing. EMBO J. 1999;18:4560–4570. doi: 10.1093/emboj/18.16.4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Le Hir H, Izaurralde E, Maquat LE, Moore MJ. The spliceosome deposits multiple proteins 20-24 nucleotides upstream of mRNA exon-exon junctions. EMBO J. 2000;19:6860–6869. doi: 10.1093/emboj/19.24.6860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chan CC, Dostie J, Diem MD, Feng W, Mann M, Rappsilber J, Dreyfuss G. eIF4A3 is a novel component of the exon junction complex. RNA. 2004;10:200–209. doi: 10.1261/rna.5230104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Swanson MS, Dreyfuss G. Classification and purification of proteins of heterogeneous nuclear ribonucleoprotein particles by RNA-binding specificities. Mol. Cell. Biol. 1988;8:2237–2241. doi: 10.1128/mcb.8.5.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baltz AG, Munschauer M, Schwanhäusser B, Vasile A, Murakawa Y, Schueler M, Youngs N, Penfold-Brown D, Drew K, Milek M, et al. The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol. Cell. 2012;46:674–690. doi: 10.1016/j.molcel.2012.05.021. [DOI] [PubMed] [Google Scholar]
- 38.Yik JH, Chen R, Nishimura R, Jennings JL, Link AJ, Zhou Q. Inhibition of P-TEFb (CDK9/Cyclin T) kinase and RNA polymerase II transcription by the coordinated actions of HEXIM1 and 7SK snRNA. Mol. Cell. 2003;12:971–982. doi: 10.1016/s1097-2765(03)00388-5. [DOI] [PubMed] [Google Scholar]
- 39.Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SRsplicing factor phosphorylation. Mol. Cell. 2010;39:925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jurica MS, Licklider LJ, Gygi SR, Grigorieff N, Moore MJ. Purification and characterization of native spliceosomes suitable for three-dimensional structural analysis. RNA. 2002;4:426–439. doi: 10.1017/s1355838202021088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bessonov S, Anokhina M, Will CL, Urlaub H, Luhrmann R. Isolation of an active step I spliceosome and composition of its RNP core. Nature. 2008;452:846–850. doi: 10.1038/nature06842. [DOI] [PubMed] [Google Scholar]
- 42.Bessonov S, Anokhina M, Krasauskas A, Golas MM, Sander B, Will CL, Urlaub H, Stark H, Lührmann R. Characterization of purified human Bact spliceosomal complexes reveals compositional and morphological changes during spliceosome activation and first step catalysis. RNA. 2010;12:2384–2403. doi: 10.1261/rna.2456210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ilagan JO, Chalkley RJ, Burlingame AL, Jurica MS. Rearrangements within human spliceosomes captured after exon ligation. RNA. 2013;3:400–412. doi: 10.1261/rna.034223.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fu XD. The superfamily of arginine/serine-rich splicing factors. RNA. 1995;1:663–680. [PMC free article] [PubMed] [Google Scholar]
- 45.Eilbracht J, Schmidt-Zachmann MS. Identification of a sequence element directing a protein to nuclear speckles. Proc. Natl Acad. Sci. USA. 2001;98:3849–3854. doi: 10.1073/pnas.071042298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jagiello I, Van Eynde A, Vulsteke V, Beullens M, Boudrez A, Keppens S, Stalmans W, Bollen M. Nuclear and subnuclear targeting sequences of the protein phosphatase-1 regulator NIPP1. J. Cell Sci. 2000;113:3761–3768. doi: 10.1242/jcs.113.21.3761. [DOI] [PubMed] [Google Scholar]
- 47.Salichs E, Ledda A, Mularoni L, Albà MM, de la Luna S. Genome-wide analysis of histidine repeats reveals their role in the localization of human proteins to the nuclear speckles compartment. PLoS Genet. 2009;5:e1000397. doi: 10.1371/journal.pgen.1000397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Loyer P, Trembley JH, Lahti JM, Kidd VJ. The RNP protein, RNPS1, associates with specific isoforms of the p34cdc2-related PITSLRE protein kinase in vivo. J. Cell Sci. 1998;111:1495–1506. doi: 10.1242/jcs.111.11.1495. [DOI] [PubMed] [Google Scholar]
- 49.Sharma A, Takata H, Shibahara K, Bubulya A, Bubulya PA. Son is essential for nuclear speckle organization and cell cycle progression. Mol. Biol. Cell. 2010;21:650–663. doi: 10.1091/mbc.E09-02-0126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Daguenet E, Baguet A, Degot S, Schmidt U, Alpy F, Wendling C, Spiegelhalter C, Kessler P, Rio MC, Le Hir H, et al. Perispeckles are major assembly sites for the exon junction core complex. Mol. Biol. Cell. 2012;9:1765–1782. doi: 10.1091/mbc.E12-01-0040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou Z, Licklider LJ, Gygi SP, Reed R. Comprehensive proteomic analysis of the human spliceosome. Nature. 2002;419:182–185. doi: 10.1038/nature01031. [DOI] [PubMed] [Google Scholar]
- 52.Hartmuth K, Urlaub H, Vornlocher HP, Will CL, Gentzel M, Wilm M, Lührmann R. Protein composition of human prespliceosomes isolated by a tobramycin affinity-selection method. Proc. Natl Acad. Sci. USA. 2002;99:719–724. doi: 10.1073/pnas.262483899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kameoka S, Duque P, Konarska MM. p54nrb associates with the 5′ splice site within large transcription/splicing complexes. EMBO J. 2004;23:1782–1791. doi: 10.1038/sj.emboj.7600187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu S, Green MR. Identification of a human protein that recognizes the 3′ splice site during the second step of pre-mRNA splicing. EMBO J. 1997;16:4421–4432. doi: 10.1093/emboj/16.14.4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Meissner M, Lopato S, Gotzmann J, Sauermann G, Barta A. Proto-oncoprotein TLS/FUS is associated to the nuclear matrix and complexed with splicing factors PTB, SRm160, and SR proteins. Exp. Cell. Res. 2003;283:184–195. doi: 10.1016/s0014-4827(02)00046-0. [DOI] [PubMed] [Google Scholar]
- 56.Chansky HA, Hu M, Hickstein DD, Yang L. Oncogenic TLS/ERG and EWS/Fli-1 fusion proteins inhibit RNA splicing mediated by YB-1 protein. Cancer Res. 2001;61:3586–3590. [PubMed] [Google Scholar]
- 57.Rapp TB, Yang L, Conrad EU, Mandahl N, Chansky HA. RNA splicing mediated by YB-1 is inhibited by TLS/CHOP in human myxoid liposarcoma cells. J. Orthop. Res. 2002;20:723–729. doi: 10.1016/S0736-0266(02)00006-2. [DOI] [PubMed] [Google Scholar]
- 58.Goransson M, Wedin M, Aman P. Temperature-dependent localization of TLS CHOP to splicing factor compartments. Exp. Cell Res. 2002;278:125–132. doi: 10.1006/excr.2002.5566. [DOI] [PubMed] [Google Scholar]
- 59.May WA, Lessnick SL, Braun BS, Klemsz M, Lewis BC, Lunsford LB, Hromas R, Denny CT. The Ewing’s sarcoma EWS/FLI–1 fusion gene encodes a more potent transcriptional activator and is a more powerful transforming gene than FLI–1. Mol. Cell. Biol. 1993;13:7393–7398. doi: 10.1128/mcb.13.12.7393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zinszner H, Albalat R, Ron D. A novel effector domain from the RNA–binding protein TLS or EWS is required for oncogenic transformation by CHOP. Genes Dev. 1994;8:2513–2526. doi: 10.1101/gad.8.21.2513. [DOI] [PubMed] [Google Scholar]
- 61.Lerga A, Hallier M, Delva L, Orvain C, Gallais I, Marie J, Moreau-Gachelin F. Identification of an RNA binding specificity for the potential splicing factor TLS. J. Biol. Chem. 2001;276:6807–6816. doi: 10.1074/jbc.M008304200. [DOI] [PubMed] [Google Scholar]
- 62.Kiledjian M, Dreyfuss G. Primary structure and binding activity of the hnRNP U protein: binding RNA through RGG box. EMBO J. 1992;11:2655–2664. doi: 10.1002/j.1460-2075.1992.tb05331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Darnell JC, Jensen KB, Jin P, Brown V, Warren ST, Darnell RB. Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell. 2001;107:489–499. doi: 10.1016/s0092-8674(01)00566-9. [DOI] [PubMed] [Google Scholar]
- 64.Ohno T, Ouchida M, Lee L, Gatalica Z, Rao VN, Reddy ES. The EWS gene, involved in Ewing family of tumors, malignant melanoma of soft parts and desmoplastic small round cell tumors, codes for an RNA binding protein with novel regulatory domains. Oncogene. 1994;9:3087–3097. [PubMed] [Google Scholar]
- 65.Burd CG, Dreyfuss G. Conserved structures and diversity of functions of RNA–binding proteins. Science. 1994;265:615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- 66.Yang L, Embree LJ, Tsai S, Hickstein DD. Oncoprotein TLS interacts with serine–arginine proteins involved in RNA splicing. J. Biol. Chem. 1998;273:27761–27764. doi: 10.1074/jbc.273.43.27761. [DOI] [PubMed] [Google Scholar]
- 67.Blackwell E, Zhang X, Ceman S. Arginines of the RGG box regulate FMRP association with polyribosomes and mRNA. Hum. Mol. Genet. 2010;19:1530–1539. doi: 10.1093/hmg/ddq007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Iko Y, Kodama S, Kasai N, Oyama T, Morita EH, Muto T, Okumura M, Fujii R, Takumi T, Tate S, et al. Domain architectures and characterization of an RNA-binding protein, TLS. J. Biol. Chem. 2004;279:44834–44840. doi: 10.1074/jbc.M408552200. [DOI] [PubMed] [Google Scholar]
- 69.Sharma S, Kohlstaedt LA, Damianov A, Rio DC, Black DL. Polypyrimidine tract binding protein controls the transition from exon definition to an intron defined spliceosome. Nat. Struct. Mol. Biol. 2008;2:183–191. doi: 10.1038/nsmb.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Behzadnia N, Golas MM, Hartmuth K, Sander B, Kastner B, Deckert J, Dube P, Will CL, Urlaub H, Stark H, et al. Composition and three-dimensional EM structure of double affinity-purified, human prespliceosomal A complexes. EMBO J. 2007;6:1737–1748. doi: 10.1038/sj.emboj.7601631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Deckert J, Hartmuth K, Boehringer D, Behzadnia N, Will CL, Kastner B, Stark H, Urlaub H, Lührmann R. Protein composition and electron microscopy structure of affinity-purified human spliceosomal B complexes isolated under physiological conditions. Mol. Cell. Biol. 2006;14:5528–5543. doi: 10.1128/MCB.00582-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Makarov EM, Owen N, Bottrill A, Makarova OV. Functional mammalian spliceosomal complex E contains SMN complex proteins in addition to U1 and U2 snRNPs. Nucleic Acids Res. 2012;6:2639–2652. doi: 10.1093/nar/gkr1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ishigaki S, Masuda A, Fujioka Y, Iguchi Y, Katsuno M, Shibata A, Urano F, Sobue G, Ohno K. Position-dependent FUS-RNA interactions regulate alternative splicing events and transcriptions. Sci. Rep. 2012;2:529. doi: 10.1038/srep00529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vance C, Rogelj B, Hortobagyi T, De Vos KJ, Nishimura AL, Sreedharan J, Hu X, Smith B, Ruddy DM, Wright P, et al. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009;323:1208–1211. doi: 10.1126/science.1165942. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.