Figure 1.
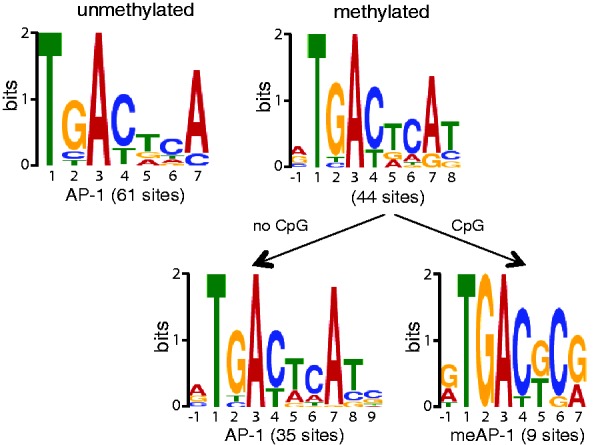
Bioinformatic identification of c-Jun/c-Fos binding motifs in the EBV genome. Deep sequencing data were analyzed with the SISSRs algorithms for putative stretches of DNA, which c-Jun/c-Fos binds. The output files of SISSRs were used as training sets for MEME, which identifies gapless, local and multiple sequence motifs. A total of 61 motifs were identified in the DNA free of CpG methylation and 44 motifs were identified in the fully CpG-methylated EBV DNA. The identified motifs in methylated DNA were selected at the level of the SISSRs training set data and grouped into motifs with and without CpG dinucleotides followed by MEME analysis.
