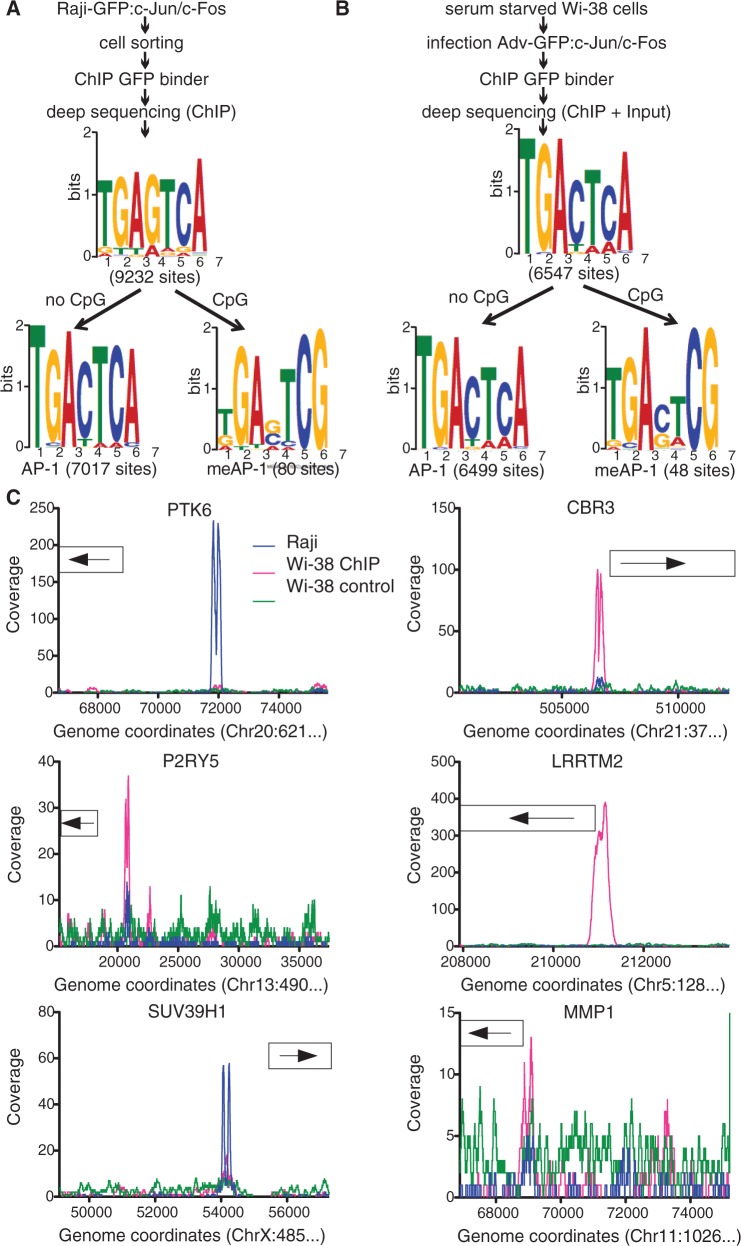
Figure 3.
Identification of AP-1 binding motifs in the human genome. Identification of c-Jun/c-Fos binding motifs in Raji (A) and Wi-38 (B) cells. The samples used for deep sequencing were prepared as indicated. Data obtained from Raji and Wi-38 cell ChIPs were analyzed with the algorithms QuEST and SISSRs, respectively. MEME analysis was conducted as described in detail in the legend of Figure 1. In chromatin of Raji and Wi-38 cells, 9232 and 6547 motifs, respectively, were discovered, which were subsequently split into two groups. One group contains all motifs identified with the aid of the MEME algorithm harboring a CpG pair in positions 6 and 7 of the consensus motif. In all, 80 and 48 motifs were identified in Raji and Wi-38 cells, respectively, termed meAP-1. The remaining motifs were reanalyzed by MEME, which identified 7017 and 6499 sites that correspond to the known consensus AP-1 site lacking a CpG dinucleotide. (C) Visualization of selected human promoters with meAP-1 sites that bind c-Jun/c-Fos in vivo. The ‘collagenase’ (MMP1) promoter served as a positive control. Depicted is the read depth (coverage) at single base pair resolution (y-axis). The boxes indicate the position of the transcripts. Genomic coordinates correspond to the human genome assembly GRCh37/hg19.