Table 2.
Data collection and refinement statistics
| Se-HpDprA(5-225) | HpDprA(5-225) | HpDprA(5-217)–ssDNA | |
|---|---|---|---|
| Data collection | |||
| Space group | P21 | P21212 | P21 |
| Cell dimensions | |||
| a, b, c (Å) | 117.88, 42.43, 181.33 | 171.09, 107.86, 37.47 | 37.40, 40.51, 154.68 |
| α, β, γ (°) | 90, 108.65, 90 | 90, 90, 90 | 90, 92.18, 90 |
| Peak | |||
| Wavelength (Å) | 0.9789 | 0.9792 | 1.0000 |
| Resolution (Å) | 59.23-2.35 (2.48-2.35) | 45.62-2.20 (2.32-2.20) | 40.51-1.80 (1.90-1.80) |
| Unique reflections | |||
| Rmergea (%) | 7.4 (24.8) | 8.6 (22.7) | 6.3 (31.1) |
| I/σ | 14.7 (6.0) | 17.9 (7.8) | 9.8 (2.9) |
| Completeness (%) | 99.1 (97.3) | 95.9 (80.9) | 83.6 (77.0) |
| Redundancy | 6.7 (6.2) | 11.1 (8.4) | 3.3 (3.5) |
| Refinement | |||
| Resolution (Å) | 39.77-2.20 | 33.16-1.80 | |
| No. of reflections | 34 008 | 34 988 | |
| Rwork/Rfreeb (%) | 16.96/21.30 | 19.40/23.71 | |
| No. of atoms | |||
| Protein | 3440 | 3318 | |
| ssDNA | 142 | ||
| Water | 321 | 385 | |
| B-factors (Å2) | |||
| Protein | 36.525 | 30.502 | |
| ssDNA | 51.915 | ||
| Water | 42.673 | 40.080 | |
| R.m.s.d. | |||
| Bond lengths (Å) | 0.0060 | 0.0040 | |
| Bond angles (°) | 0.976 | 0.870 |
Values in parentheses indicate the specific values in the highest resolution shell.
aRmerge =  , where
, where  is the intensity of the measured reflection, and
is the intensity of the measured reflection, and 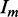 is the mean intensity of all the symmetry-related reflections.
is the mean intensity of all the symmetry-related reflections.
bRwork =  , where
, where 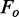 denotes the observed structure factor amplitude and
denotes the observed structure factor amplitude and 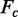 denotes the structure factor amplitude calculated from the model. Rfree is as for Rwork but calculated with 5.0% of randomly chosen reflections omitted from the refinement.
denotes the structure factor amplitude calculated from the model. Rfree is as for Rwork but calculated with 5.0% of randomly chosen reflections omitted from the refinement.
