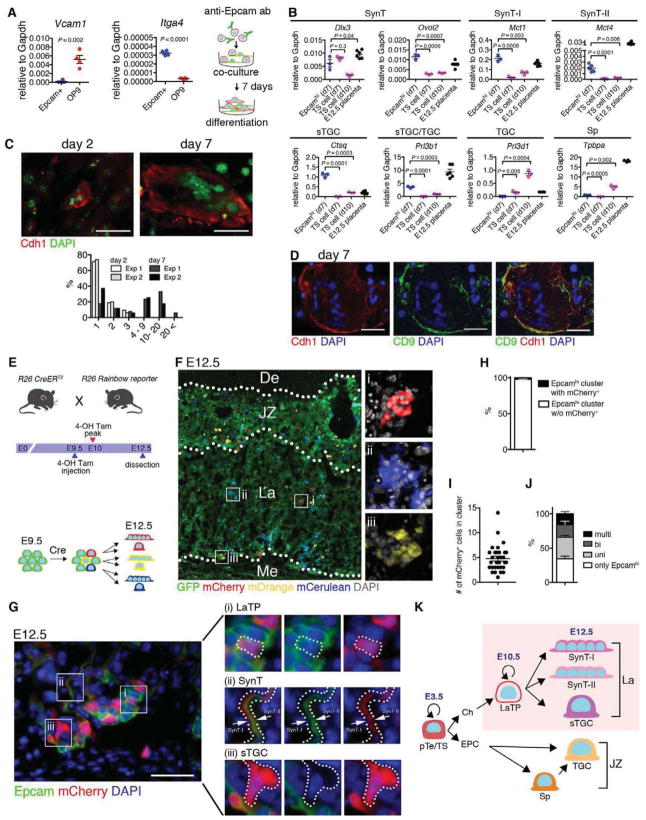
Figure 2. Epcamhi cells represent multipotent labyrinth trophoblast progenitors.
(A) Schematic for analysis of the developmental potential of Epcamhi cells in vitro. Epcamhi cells from E10.5 placenta were enriched using magnetic beads and co-cultured with OP9 for 7 days. QRT-PCR documents the expression of Itga4 in Epcamhi cells and Vcam1 in OP9.
(B) QRT-PCR showing the maintenance of SynT specific genes and up-regulation of markers for differentiated SynT-I and -II and sTGC in Epcamhi cultures after 7 days.
(C) Documentation of increased frequency of Cdh1+ SynT with multiple nuclei after 7 days in culture.
(D) Documentation that Cdh1+ multinucleated cells co-express trophoblast marker CD9.
(E) Schematic for in vivo clonality analysis. Rosa26 (R26) Rainbow reporter mice were mated with R26 CreERT2 mice. At E9.5, Cre-mediated gene deletion was induced by injection with 4OH-tamoxifen, and embryos were dissected at E12.5.
(F) Fluorescence image indicating that Cre-mediated gene recombination induces multi-color labeling and establishment of clones of labeled cells in the labyrinth. JZ, junctional zone. La, labyrinth. Me, mesenchyme.
(G) Documentation of multi-lineage differentiation in a cluster of mCherry+ labeled trophoblasts. (i) Epcamhi (LaTP), (ii) SynT (Epcamlow/neg) and (iii) sTGC (Epcamneg with large nuclei).
(H) Analysis of the frequency of clusters containing mCherry-labeled Epcamhi cells documenting infrequent labeling of clusters with same fluorescent color.
(I) Average number of mCherry+ cells in a labeled cluster documenting the establishment of multicellular, labeled clones.
(J) Differentiation potential of mCherry-labeled cells documenting the presence of clones with Epcamhi LaTP and all labyrinth trophoblast subtypes.
(K) Schematic representing the hierarchy of trophoblast-lineage differentiation. LaTP gives rise to all types of labyrinth trophoblasts, but not Sp or TGC.
All error bars indicate SEM (Standard error of mean).
See also Figure S2.