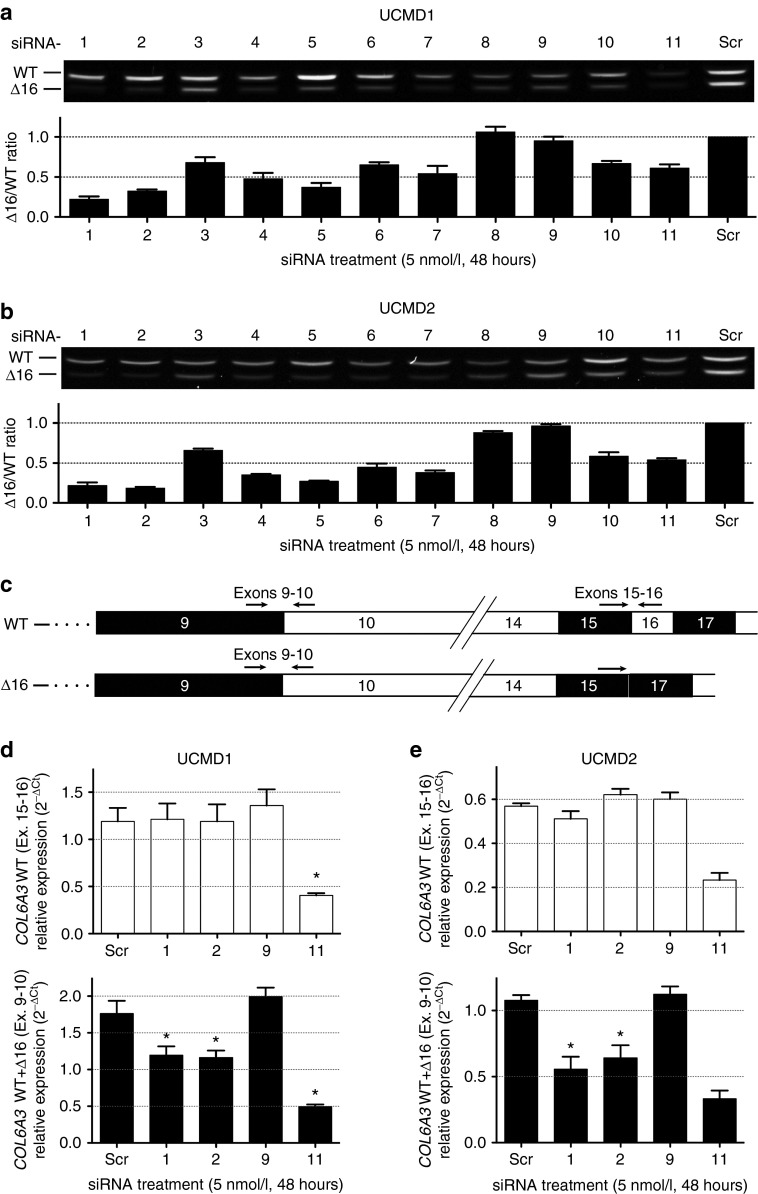
Figure 2.
In vitro silencing of mutant COL6A3 mRNA transcripts (Δ16) is achieved by allele-specific small interfering RNA (siRNA) oligos in two congenital muscular dystrophy type Ullrich (UCMD) fibroblast lines. (a,b) Agarose gel representing a single transfection (48 hours) of each siRNA (including a scrambled control – Scr) into two UCMD fibroblast cell lines (UCMD1, a; UCMD2, b). Semiquantitative RT-PCR was performed using primers in exons 14 and 20, and was ended after 26 cycles, to prevent saturation. The lower band on the gels represents the mutant transcript (Δ16). The intensity of each fragment was quantified using ImageJ, and the allele-specific knockdown was reported as the intensity ratio of the mutant over the wild-type (Δ16/WT) fragments (graphs shown below each gel image). Each bar on the graphs represents the average of three single transfections (amplified three times each) ± standard error of the mean (SEM). (c) Schematic of the COL6A3 mRNA from the wild-type (WT) and the mutant (Δ16) alleles, showing the position of the primers used for the quantitative RT-PCR (qRT-PCR) experiment. (d,e) The relative expression of only the WT COL6A3 transcripts (that include exon 16, primers placed in exons 15 and 16, upper panels) or of the total COL6A3 transcripts (primers placed in exons 9 and 10, lower panels) was measured by qRT-PCR, using β-actin (ACTB) and phosphoglycerate kinase 1 (PGK1) as endogenous controls, in fibroblasts from (d) UCMD1 and (e) UCMD2 following transfection of siRNA oligos. Graphs depict the 2−ΔCt data. Each bar is the average of three single transfections (each amplified in triplicate) ± SEM. *P ≤ 0.05, one-tailed Mann–Whitney. For UCMD2, comparison of scrambled- versus siRNA-11 treatments could not be completed (n = 2 for these datapoints).