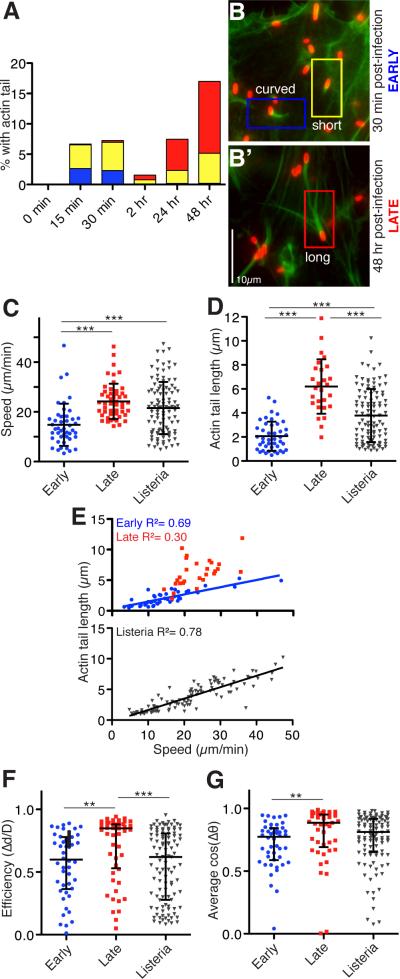
Figure 1. R. parkeri motility occurs in early and late phases with distinct movement characteristics.
(A) Graph depicting the percentage of R. parkeri with actin tails that are curved (blue; > 90° bend in tail), short (yellow, < 2 bacteria lengths) or long (red, > 2 bacteria lengths) in HMEC-1 cells infected synchronously for the indicated times as described previously [14]. Results are the average from three independent experiments performed in duplicate. (B) R. parkeri infected cells were fixed at the indicated times, stained with anti-Rickettsia antibody (red) and for actin with Alexa Fluor 488-phalloidin (green). (C-G) Bacterial motility parameters in HMEC-1 cells expressing Lifeact-EGFP and infected with R. parkeri expressing mCherry [15] early (12-60 mpi) or late (48 hpi), or HMEC-1 cells expressing Lifeact-mCherry and infected with L. monocytogenes strain 10403S expressing GFP [16] at 8-12 hpi. (C) Speed of movement and (D) actin tail length for R. parkeri early (blue) and late (red), and for L. monocytogenes (grey). Mean ± SD, *** p<0.001 by ANOVA with Bonferroni's Multiple Comparison Test, from three separate experiments, with tracking and speed data in 60 s intervals for each bacterium. (E) Relationship between average speed and actin tail length for each bacterium, with best-fit linear regression. (F) Average efficiency of movement, calculated by dividing the total x-y displacement by the total distance moved over 60 s for each bacterium. (G) Path straightness for each bacterium, calculated by averaging cosines of the change in tangent angle between adjacent track segments (Δθ) over 60 s of movement. For (F, G), we analyzed variation around the medians due to the non-Gaussian distribution of the data. Data are median ± interquartile range, ** p<0.01, *** p<0.001 by Kruskal-Wallis test with Dunn's Multiple Comparison test. See also Figure S1, Movies S1-S3.