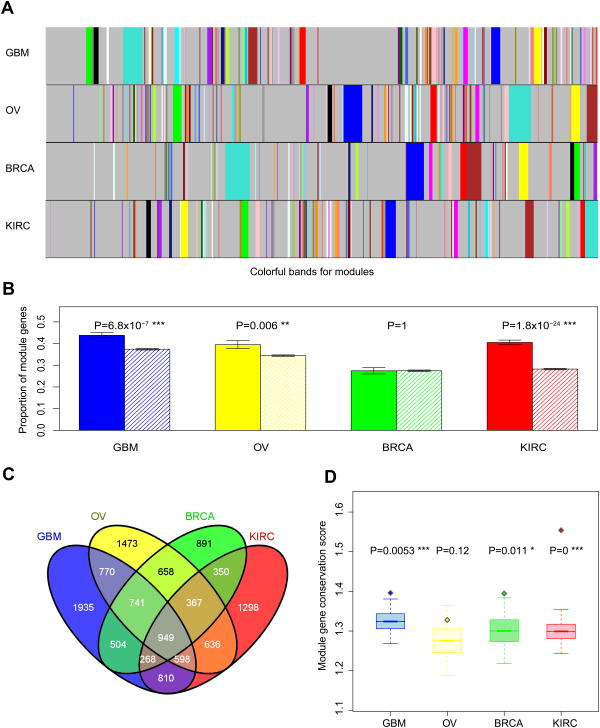
Figure 2. The enrichment of prognostic mRNA genes in modules.
(A) Modules defined from the weighted gene co-expression networks. Colorful bands represent modules in the network, with the biggest module in turquoise, second largest in blue, then brown, green, yellow and so on. (B) Prognostic genes are enriched in the modules. Solid bars represent the proportions of module genes among prognostic mRNA genes; striped bars represent the proportions of module genes among non-prognostic mRNA genes. Error bars indicate ± 1 s.e.m., and P-values were calculated based on Fisher's exact tests. (C) The Venn diagram of module genes across the four cancer types. (D) Boxplots (median ± 1 quartile) showing that prognostic genes tend to be more conserved module genes. Y-axis represents the module-gene conservation score, which ranges from 0 to 4, with 0 indicating not a module gene in any of the four cancer types and 4 indicating a module gene in all four cancer types. Each boxplot represents the mean-conservation-score distribution of 20,000 randomly sampled same-size gene sets; the diamond dot represents the mean conservation score of prognostic genes. P-values were calculated based on Wilcoxon rank sum tests.