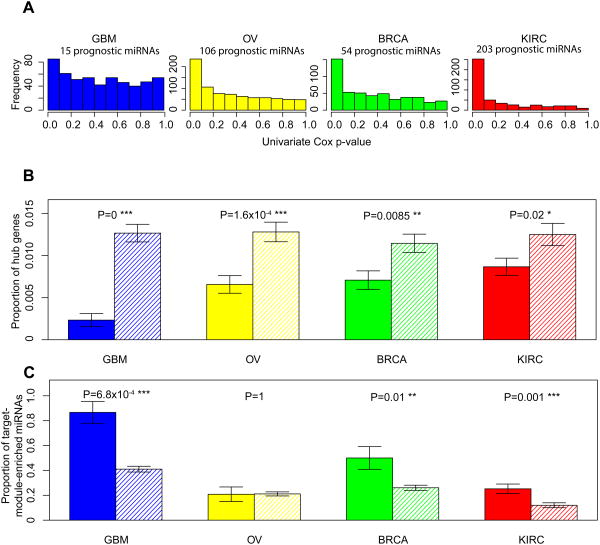
Figure 3. Target genes of prognostic miRNA genes show the same patterns.
(A) The P-value distributions of the correlations of miRNA expression with overall survival based on the univariate Cox model in the four cancer types. Based on the signal-to-noise ratio, prognostic miRNA genes were identified. (B) Target genes of prognostic miRNA genes are depleted in the hubs. Solid bars represent the proportions of hub genes among target genes of prognostic miRNAs; striped bars represent the proportions of hub genes among non-target genes of prognostic miRNAs. (C) Target genes of prognostic miRNAs are enriched in the modules. Solid bars represent the proportions of target-module–enriched miRNAs among prognostic miRNAs; striped bars represent the proportions of target-module–enriched miRNAs among non-prognostic miRNAs. Error bars indicate ± 1 s.e.m., and P-values were calculated based on Fisher's exact tests.