Table 1. Compilation of results.
| cmc | zeff | Kins/105 | Keff/108 |
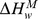
|
MIC (L. lactis) | MIC (E. coli) | H5 | |
| MPX | >1000 | 2.5±0.1 | 4.2±0.2 | 1.6±0.2 | −24.0±0.3 | 2.5±0.5 | 8±3 | 18±2 |
| Ala1 | >1000 | 4.3±0.1 | 1.1±0.1 | 25±7 | −18.4±0.3 | 18±7 | 33±8 | 230±10 |
| Ala14 | >1000 | 2.9±0.1 | 1.4±0.3 | 1.1±0.2 | −16.2±0.3 | 21±4 | 33±8 | 220±20 |
| PAMPX | >1000 | ----------- | ----------- | ----------- | ----------- | 3.3±0.8 | 21±4 | 25±1 |
| Leu8 | >1000 | ----------- | ----------- | ----------- | ----------- | 2.5±1.3 | 7±3 | 14±2 |
| Adec1 | 90±10 | ----------- | ----------- | ----------- | ----------- | 1.7±0.4 | 3.3±0.8 | 0.7±0.1 |
| Adec8 | 30±7 | ----------- | ----------- | ----------- | ----------- | 4.2±0.8 | 4.2±0.8 | 1±1 |
| Adec14 | 10±20 | ----------- | ----------- | ----------- | ----------- | 1.7±0.4 | 3.3±0.8 | 0.8±0.2 |
| OAMPX | 150±50 | ----------- | ----------- | ----------- | ----------- | 3.3±0.8 | 5±1 | 1.1±0.3 |
Critical micelle concentration (cmc in µM units), effective charge (zeff) of the peptide, partition coefficient of insertion (Kins), effective partition coefficient (Keff), molar enthalpy of partitioning (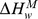 in kJ/mol units), minimal inhibitory concentration (MIC in µM units) and the haemolytic potency is given as the concentration H5 (in µM units) at which 5% haemolysis has been obtained. Membrane partitioning data for PAMPX, Leu8, Adec1, Adec8, Adec14 and OAMPX could not be analysed and these cells are marked with a dashed line.
in kJ/mol units), minimal inhibitory concentration (MIC in µM units) and the haemolytic potency is given as the concentration H5 (in µM units) at which 5% haemolysis has been obtained. Membrane partitioning data for PAMPX, Leu8, Adec1, Adec8, Adec14 and OAMPX could not be analysed and these cells are marked with a dashed line.
