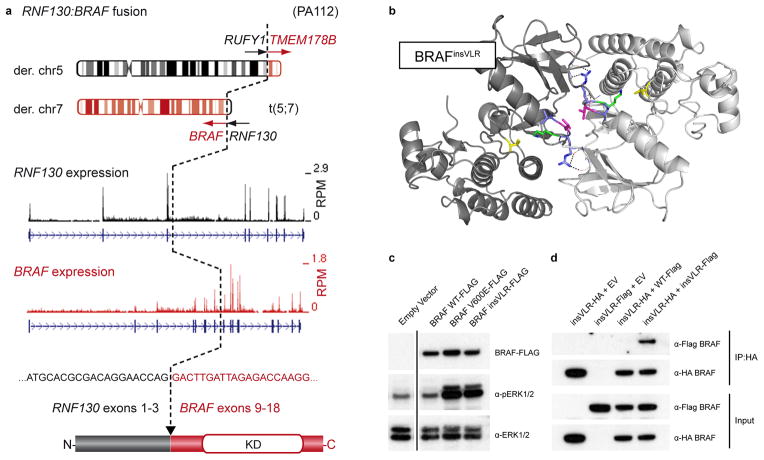
Figure 1. Novel BRAF alterations in pilocytic astrocytoma.
a, Schematic representation of the RNF130:BRAF fusion gene in ICGC_PA112 resulting from a translocation between chromosomes 5 and 7. A similar fusion was observed in ICGC_PA96. The cDNA sequence at the fusion breakpoint and resulting exon and protein structures are indicated. A reciprocal fusion between RUN and FYVE domain containing 1 (RUFY1) and transmembrane protein 178B (TMEM178B) on the derivative chromosome 5 in ICGC_PA112 was also found to be expressed by RNA-seq analysis (not shown). RPM, reads per million; KD, kinase domain.
b, Computational modeling of two BRAF monomers (light/dark grey) with a VLR insertion (blue/magenta) between Arg506 and Lys507 (green), as identified in ICGC_PA65. The PDB structure 4E26 was used as a template. Val600, a mutational hotspot, is shown in yellow. A novel dimer interface is formed between the protomers, with hydrogen bonds from the new arginine side chains (dashed lines) and a hydrophobic interaction between the leucine side chains (magenta).
c, Western blot analysis of NIH3T3 cells transfected with empty vector (EV), BRAFWT, BRAFV600E and BRAFinsVLR. The newly identified insVLR mutant results in elevation of ERK1/2 phosphorylation (pERK1/2) at a similar level to that seen with the known oncogenic V600E form.
d, Co-immunoprecipation assay with pull-down of HA-tagged BRAFinsVLR, demonstrating that this novel mutant forms homodimers with co-transfected AU1-tagged insVLR, but does not appear to form a strong heterodimer with wild-type BRAF.