Abstract
Human immunodeficiency virus type 1 (HIV-1) RNA contains an extended hairpin structure at the 5' end (the TAR element) that is essential for viral replication. The upper part of the stem-loop structure binds the virally encoded transcriptional activator protein Tat and cellular co-factors, but no clear function for the lower stem region has been established. Here, we report that mutant HIV-1 viruses with base substitutions in the lower stem region are dead, most likely at the level of transcription from an integrated provirus. By using large amounts of these mutant DNA constructs for transfections, revertant viruses with a great variety of genetic changes (point mutations, short deletions) could be isolated in prolonged culture experiments that lasted over 6 months. The pattern and evolution of these changes supported the notion that base-pairing of the lower stem region is essential for optimal HIV-1 replication. The functional and genetic plasticities of this RNA domain and the HIV-1 long terminal repeat promoter are discussed.
Full text
PDF

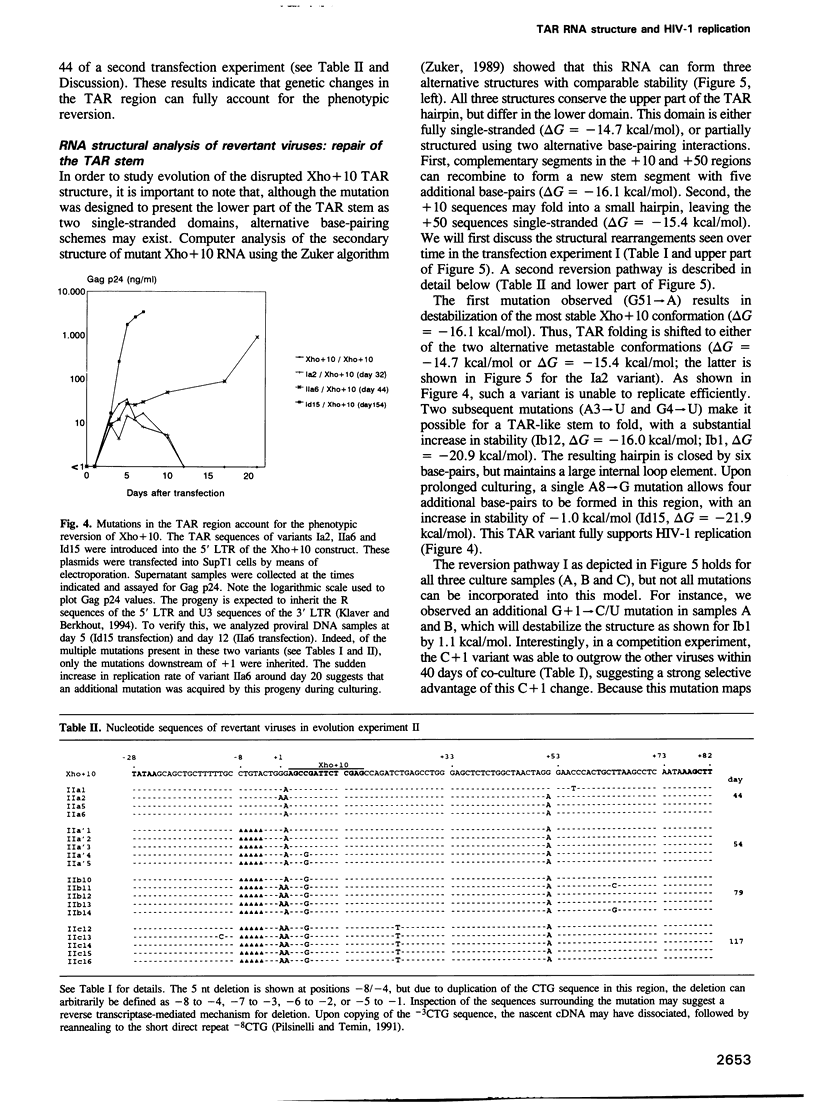
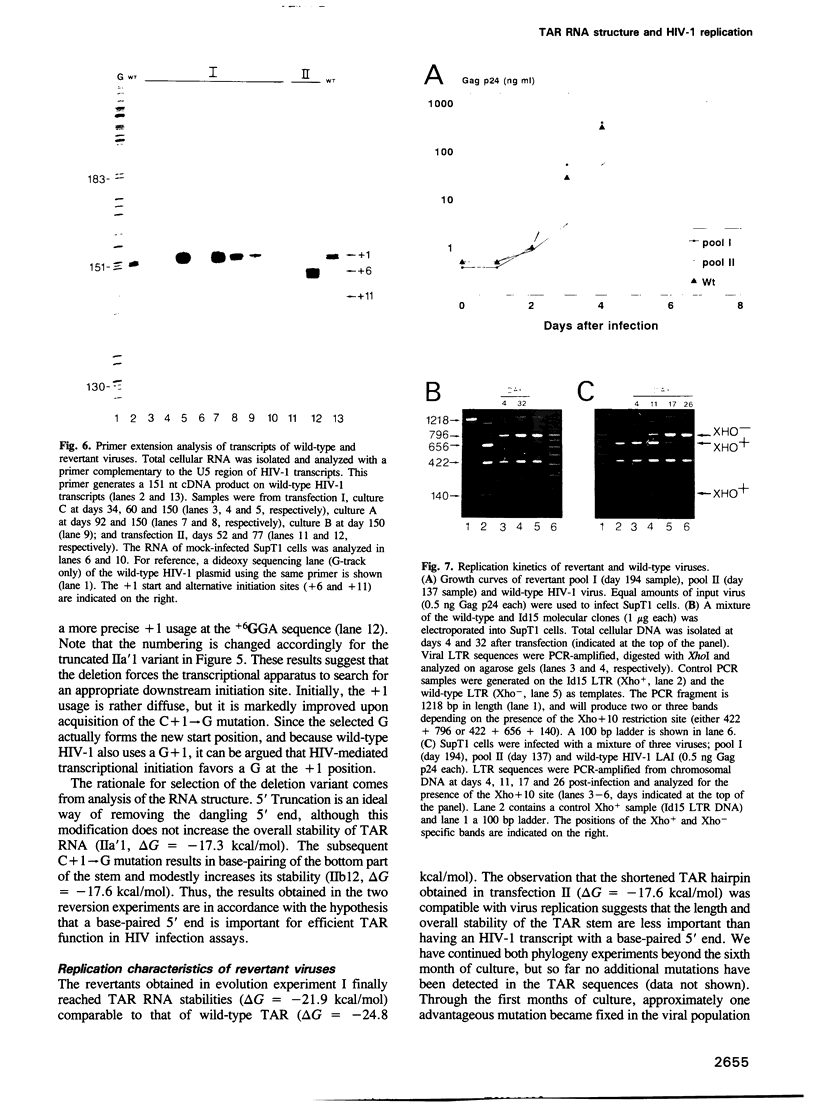
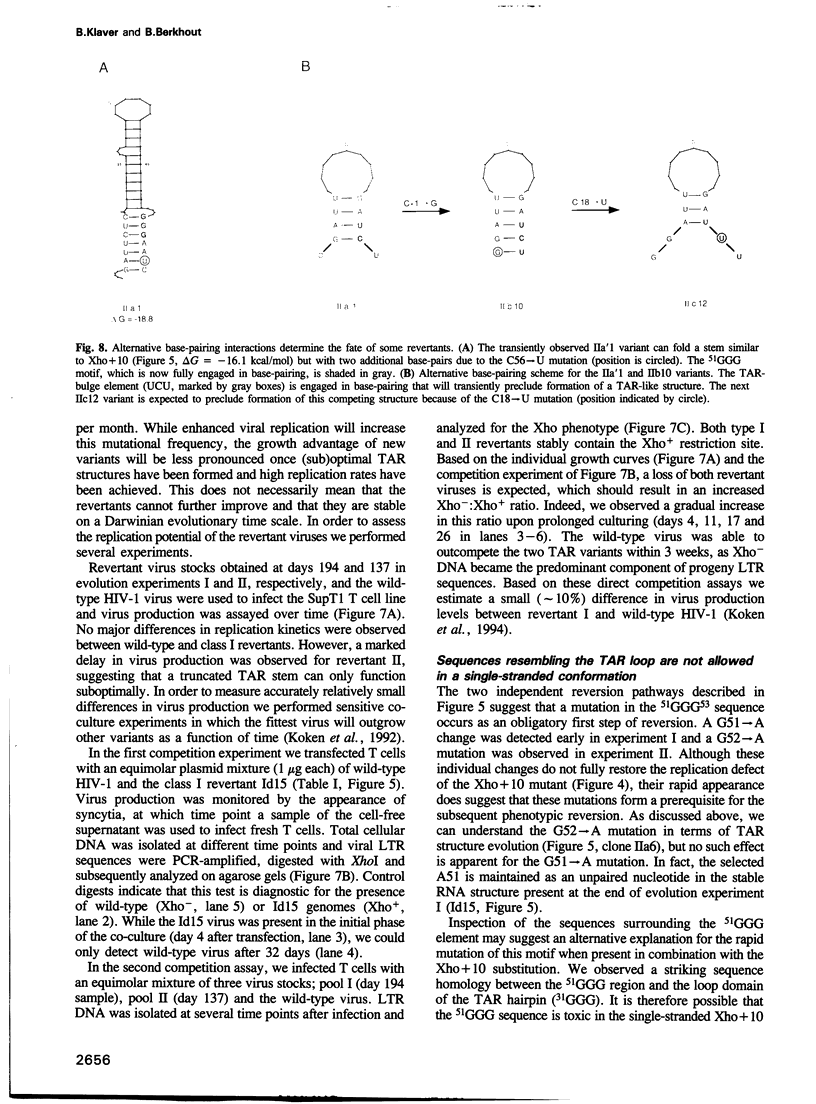



Images in this article
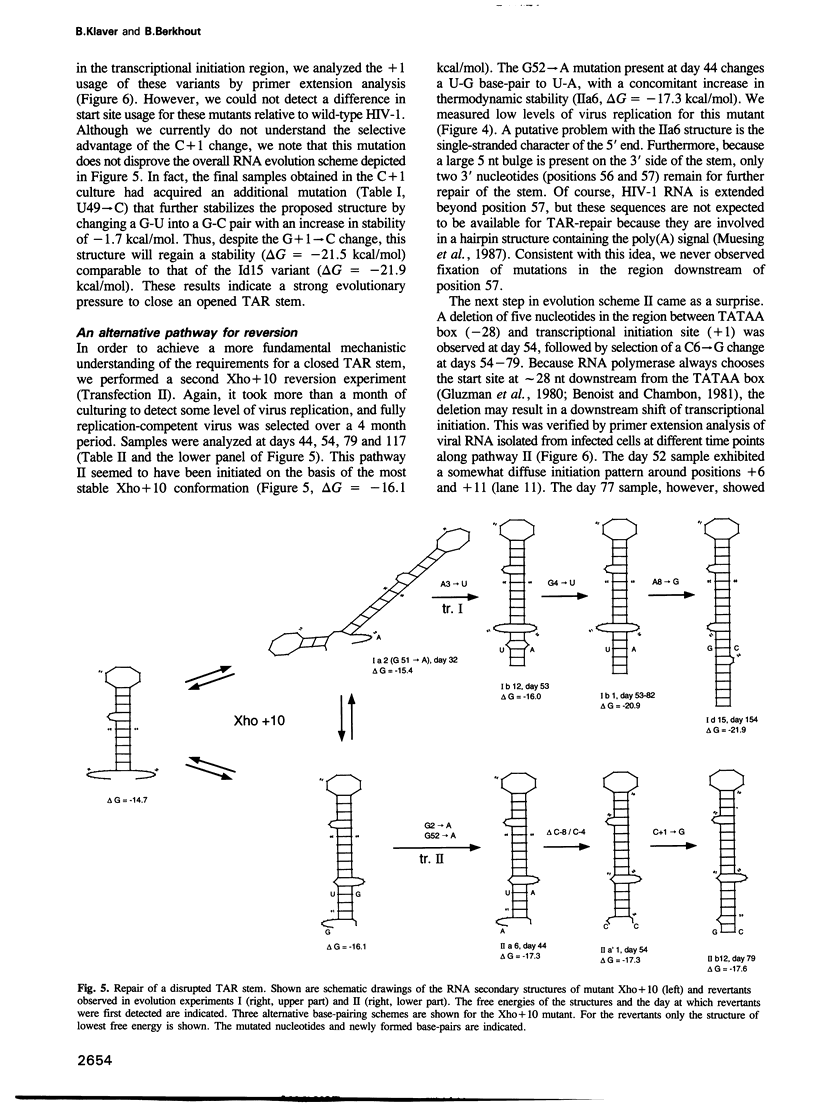
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams C. C., Workman J. L. Nucleosome displacement in transcription. Cell. 1993 Feb 12;72(3):305–308. doi: 10.1016/0092-8674(93)90109-4. [DOI] [PubMed] [Google Scholar]
- Bagasra O., Khalili K., Seshamma T., Taylor J. P., Pomerantz R. J. TAR-independent replication of human immunodeficiency virus type 1 in glial cells. J Virol. 1992 Dec;66(12):7522–7528. doi: 10.1128/jvi.66.12.7522-7528.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudin F., Marquet R., Isel C., Darlix J. L., Ehresmann B., Ehresmann C. Functional sites in the 5' region of human immunodeficiency virus type 1 RNA form defined structural domains. J Mol Biol. 1993 Jan 20;229(2):382–397. doi: 10.1006/jmbi.1993.1041. [DOI] [PubMed] [Google Scholar]
- Benoist C., Chambon P. In vivo sequence requirements of the SV40 early promotor region. Nature. 1981 Mar 26;290(5804):304–310. doi: 10.1038/290304a0. [DOI] [PubMed] [Google Scholar]
- Berkhout B., Gatignol A., Silver J., Jeang K. T. Efficient trans-activation by the HIV-2 Tat protein requires a duplicated TAR RNA structure. Nucleic Acids Res. 1990 Apr 11;18(7):1839–1846. doi: 10.1093/nar/18.7.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkhout B., Jeang K. T. Detailed mutational analysis of TAR RNA: critical spacing between the bulge and loop recognition domains. Nucleic Acids Res. 1991 Nov 25;19(22):6169–6176. doi: 10.1093/nar/19.22.6169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkhout B., Jeang K. T. trans activation of human immunodeficiency virus type 1 is sequence specific for both the single-stranded bulge and loop of the trans-acting-responsive hairpin: a quantitative analysis. J Virol. 1989 Dec;63(12):5501–5504. doi: 10.1128/jvi.63.12.5501-5504.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkhout B., Klaver B. In vivo selection of randomly mutated retroviral genomes. Nucleic Acids Res. 1993 Nov 11;21(22):5020–5024. doi: 10.1093/nar/21.22.5020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkhout B., Silverman R. H., Jeang K. T. Tat trans-activates the human immunodeficiency virus through a nascent RNA target. Cell. 1989 Oct 20;59(2):273–282. doi: 10.1016/0092-8674(89)90289-4. [DOI] [PubMed] [Google Scholar]
- Berkhout B. Structural features in TAR RNA of human and simian immunodeficiency viruses: a phylogenetic analysis. Nucleic Acids Res. 1992 Jan 11;20(1):27–31. doi: 10.1093/nar/20.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke J. M., Berzal-Herranz A. In vitro selection and evolution of RNA: applications for catalytic RNA, molecular recognition, and drug discovery. FASEB J. 1993 Jan;7(1):106–112. doi: 10.1096/fasebj.7.1.8422956. [DOI] [PubMed] [Google Scholar]
- Churcher M. J., Lamont C., Hamy F., Dingwall C., Green S. M., Lowe A. D., Butler J. G., Gait M. J., Karn J. High affinity binding of TAR RNA by the human immunodeficiency virus type-1 tat protein requires base-pairs in the RNA stem and amino acid residues flanking the basic region. J Mol Biol. 1993 Mar 5;230(1):90–110. doi: 10.1006/jmbi.1993.1128. [DOI] [PubMed] [Google Scholar]
- Cordingley M. G., LaFemina R. L., Callahan P. L., Condra J. H., Sardana V. V., Graham D. J., Nguyen T. M., LeGrow K., Gotlib L., Schlabach A. J. Sequence-specific interaction of Tat protein and Tat peptides with the transactivation-responsive sequence element of human immunodeficiency virus type 1 in vitro. Proc Natl Acad Sci U S A. 1990 Nov;87(22):8985–8989. doi: 10.1073/pnas.87.22.8985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen B. R. Mechanism of action of regulatory proteins encoded by complex retroviruses. Microbiol Rev. 1992 Sep;56(3):375–394. doi: 10.1128/mr.56.3.375-394.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai K., Loewenstein P. M., Green M. Isolation of a cellular protein that binds to the human immunodeficiency virus Tat protein and can potentiate transactivation of the viral promoter. Proc Natl Acad Sci U S A. 1991 Oct 15;88(20):8875–8879. doi: 10.1073/pnas.88.20.8875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrov D. S., Willey R. L., Sato H., Chang L. J., Blumenthal R., Martin M. A. Quantitation of human immunodeficiency virus type 1 infection kinetics. J Virol. 1993 Apr;67(4):2182–2190. doi: 10.1128/jvi.67.4.2182-2190.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingwall C., Ernberg I., Gait M. J., Green S. M., Heaphy S., Karn J., Lowe A. D., Singh M., Skinner M. A., Valerio R. Human immunodeficiency virus 1 tat protein binds trans-activation-responsive region (TAR) RNA in vitro. Proc Natl Acad Sci U S A. 1989 Sep;86(18):6925–6929. doi: 10.1073/pnas.86.18.6925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H., Roy A. L., Roeder R. G. Human transcription factor USF stimulates transcription through the initiator elements of the HIV-1 and the Ad-ML promoters. EMBO J. 1993 Feb;12(2):501–511. doi: 10.1002/j.1460-2075.1993.tb05682.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenfeld G. Chromatin as an essential part of the transcriptional mechanism. Nature. 1992 Jan 16;355(6357):219–224. doi: 10.1038/355219a0. [DOI] [PubMed] [Google Scholar]
- Feng S., Holland E. C. HIV-1 tat trans-activation requires the loop sequence within tar. Nature. 1988 Jul 14;334(6178):165–167. doi: 10.1038/334165a0. [DOI] [PubMed] [Google Scholar]
- Frankel A. D., Biancalana S., Hudson D. Activity of synthetic peptides from the Tat protein of human immunodeficiency virus type 1. Proc Natl Acad Sci U S A. 1989 Oct;86(19):7397–7401. doi: 10.1073/pnas.86.19.7397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freier S. M., Kierzek R., Jaeger J. A., Sugimoto N., Caruthers M. H., Neilson T., Turner D. H. Improved free-energy parameters for predictions of RNA duplex stability. Proc Natl Acad Sci U S A. 1986 Dec;83(24):9373–9377. doi: 10.1073/pnas.83.24.9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia J. A., Harrich D., Soultanakis E., Wu F., Mitsuyasu R., Gaynor R. B. Human immunodeficiency virus type 1 LTR TATA and TAR region sequences required for transcriptional regulation. EMBO J. 1989 Mar;8(3):765–778. doi: 10.1002/j.1460-2075.1989.tb03437.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia J. A., Wu F. K., Mitsuyasu R., Gaynor R. B. Interactions of cellular proteins involved in the transcriptional regulation of the human immunodeficiency virus. EMBO J. 1987 Dec 1;6(12):3761–3770. doi: 10.1002/j.1460-2075.1987.tb02711.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatignol A., Buckler-White A., Berkhout B., Jeang K. T. Characterization of a human TAR RNA-binding protein that activates the HIV-1 LTR. Science. 1991 Mar 29;251(5001):1597–1600. doi: 10.1126/science.2011739. [DOI] [PubMed] [Google Scholar]
- Gilboa E., Mitra S. W., Goff S., Baltimore D. A detailed model of reverse transcription and tests of crucial aspects. Cell. 1979 Sep;18(1):93–100. doi: 10.1016/0092-8674(79)90357-x. [DOI] [PubMed] [Google Scholar]
- Gluzman Y., Sambrook J. F., Frisque R. J. Expression of early genes of origin-defective mutants of simian virus 40. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3898–3902. doi: 10.1073/pnas.77.7.3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunnery S., Green S. R., Mathews M. B. Tat-responsive region RNA of human immunodeficiency virus type 1 stimulates protein synthesis in vivo and in vitro: relationship between structure and function. Proc Natl Acad Sci U S A. 1992 Dec 1;89(23):11557–11561. doi: 10.1073/pnas.89.23.11557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunnery S., Rice A. P., Robertson H. D., Mathews M. B. Tat-responsive region RNA of human immunodeficiency virus 1 can prevent activation of the double-stranded-RNA-activated protein kinase. Proc Natl Acad Sci U S A. 1990 Nov;87(22):8687–8691. doi: 10.1073/pnas.87.22.8687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrich D., Garcia J., Mitsuyasu R., Gaynor R. TAR independent activation of the human immunodeficiency virus in phorbol ester stimulated T lymphocytes. EMBO J. 1990 Dec;9(13):4417–4423. doi: 10.1002/j.1460-2075.1990.tb07892.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauber J., Cullen B. R. Mutational analysis of the trans-activation-responsive region of the human immunodeficiency virus type I long terminal repeat. J Virol. 1988 Mar;62(3):673–679. doi: 10.1128/jvi.62.3.673-679.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobovits A., Smith D. H., Jakobovits E. B., Capon D. J. A discrete element 3' of human immunodeficiency virus 1 (HIV-1) and HIV-2 mRNA initiation sites mediates transcriptional activation by an HIV trans activator. Mol Cell Biol. 1988 Jun;8(6):2555–2561. doi: 10.1128/mcb.8.6.2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeang K. T., Berkhout B., Dropulic B. Effects of integration and replication on transcription of the HIV-1 long terminal repeat. J Biol Chem. 1993 Nov 25;268(33):24940–24949. [PubMed] [Google Scholar]
- Jeang K. T., Chun R., Lin N. H., Gatignol A., Glabe C. G., Fan H. In vitro and in vivo binding of human immunodeficiency virus type 1 Tat protein and Sp1 transcription factor. J Virol. 1993 Oct;67(10):6224–6233. doi: 10.1128/jvi.67.10.6224-6233.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones K. A., Luciw P. A., Duchange N. Structural arrangements of transcription control domains within the 5'-untranslated leader regions of the HIV-1 and HIV-2 promoters. Genes Dev. 1988 Sep;2(9):1101–1114. doi: 10.1101/gad.2.9.1101. [DOI] [PubMed] [Google Scholar]
- Kato H., Horikoshi M., Roeder R. G. Repression of HIV-1 transcription by a cellular protein. Science. 1991 Mar 22;251(5000):1476–1479. doi: 10.1126/science.2006421. [DOI] [PubMed] [Google Scholar]
- Kim J. Y., Gonzalez-Scarano F., Zeichner S. L., Alwine J. C. Replication of type 1 human immunodeficiency viruses containing linker substitution mutations in the -201 to -130 region of the long terminal repeat. J Virol. 1993 Mar;67(3):1658–1662. doi: 10.1128/jvi.67.3.1658-1662.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaver B., Berkhout B. Premature strand transfer by the HIV-1 reverse transcriptase during strong-stop DNA synthesis. Nucleic Acids Res. 1994 Jan 25;22(2):137–144. doi: 10.1093/nar/22.2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koken S. E., van Wamel J. L., Goudsmit J., Berkhout B., Geelen J. L. Natural variants of the HIV-1 long terminal repeat: analysis of promoters with duplicated DNA regulatory motifs. Virology. 1992 Dec;191(2):968–972. doi: 10.1016/0042-6822(92)90274-s. [DOI] [PubMed] [Google Scholar]
- Koken S.E.C., van Wamel J.L.B., Geelen J.L.M.C., Berkhout B. Functional Analysis of the ACTGCTGA Sequence Motif in the Human Immunodeficiency Virus Type-1 Long Terminal Repeat Promoter. J Biomed Sci. 1994 Mar;1(2):83–92. doi: 10.1007/BF02257981. [DOI] [PubMed] [Google Scholar]
- Kozak M. Translation of insulin-related polypeptides from messenger RNAs with tandemly reiterated copies of the ribosome binding site. Cell. 1983 Oct;34(3):971–978. doi: 10.1016/0092-8674(83)90554-8. [DOI] [PubMed] [Google Scholar]
- Laimins L., Holmgren-König M., Khoury G. Transcriptional "silencer" element in rat repetitive sequences associated with the rat insulin 1 gene locus. Proc Natl Acad Sci U S A. 1986 May;83(10):3151–3155. doi: 10.1073/pnas.83.10.3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard J., Parrott C., Buckler-White A. J., Turner W., Ross E. K., Martin M. A., Rabson A. B. The NF-kappa B binding sites in the human immunodeficiency virus type 1 long terminal repeat are not required for virus infectivity. J Virol. 1989 Nov;63(11):4919–4924. doi: 10.1128/jvi.63.11.4919-4924.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann D. A., Frankel A. D. Endocytosis and targeting of exogenous HIV-1 Tat protein. EMBO J. 1991 Jul;10(7):1733–1739. doi: 10.1002/j.1460-2075.1991.tb07697.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marciniak R. A., Garcia-Blanco M. A., Sharp P. A. Identification and characterization of a HeLa nuclear protein that specifically binds to the trans-activation-response (TAR) element of human immunodeficiency virus. Proc Natl Acad Sci U S A. 1990 May;87(9):3624–3628. doi: 10.1073/pnas.87.9.3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda T., Harada S. Modulation of host cell nuclear proteins that bind to HIV-1 trans-activation-responsive element RNA by phorbol ester. Virology. 1993 Feb;192(2):696–700. doi: 10.1006/viro.1993.1091. [DOI] [PubMed] [Google Scholar]
- McPherson C. E., Shim E. Y., Friedman D. S., Zaret K. S. An active tissue-specific enhancer and bound transcription factors existing in a precisely positioned nucleosomal array. Cell. 1993 Oct 22;75(2):387–398. doi: 10.1016/0092-8674(93)80079-t. [DOI] [PubMed] [Google Scholar]
- Moses A. V., Ibanez C., Gaynor R., Ghazal P., Nelson J. A. Differential role of long terminal repeat control elements for the regulation of basal and Tat-mediated transcription of the human immunodeficiency virus in stimulated and unstimulated primary human macrophages. J Virol. 1994 Jan;68(1):298–307. doi: 10.1128/jvi.68.1.298-307.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muesing M. A., Smith D. H., Capon D. J. Regulation of mRNA accumulation by a human immunodeficiency virus trans-activator protein. Cell. 1987 Feb 27;48(4):691–701. doi: 10.1016/0092-8674(87)90247-9. [DOI] [PubMed] [Google Scholar]
- Nelbock P., Dillon P. J., Perkins A., Rosen C. A. A cDNA for a protein that interacts with the human immunodeficiency virus Tat transactivator. Science. 1990 Jun 29;248(4963):1650–1653. doi: 10.1126/science.2194290. [DOI] [PubMed] [Google Scholar]
- Ohana B., Moore P. A., Ruben S. M., Southgate C. D., Green M. R., Rosen C. A. The type 1 human immunodeficiency virus Tat binding protein is a transcriptional activator belonging to an additional family of evolutionarily conserved genes. Proc Natl Acad Sci U S A. 1993 Jan 1;90(1):138–142. doi: 10.1073/pnas.90.1.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak V. K., Temin H. M. 5-Azacytidine and RNA secondary structure increase the retrovirus mutation rate. J Virol. 1992 May;66(5):3093–3100. doi: 10.1128/jvi.66.5.3093-3100.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peden K., Emerman M., Montagnier L. Changes in growth properties on passage in tissue culture of viruses derived from infectious molecular clones of HIV-1LAI, HIV-1MAL, and HIV-1ELI. Virology. 1991 Dec;185(2):661–672. doi: 10.1016/0042-6822(91)90537-l. [DOI] [PubMed] [Google Scholar]
- Pulsinelli G. A., Temin H. M. Characterization of large deletions occurring during a single round of retrovirus vector replication: novel deletion mechanism involving errors in strand transfer. J Virol. 1991 Sep;65(9):4786–4797. doi: 10.1128/jvi.65.9.4786-4797.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rounseville M. P., Kumar A. Binding of a host cell nuclear protein to the stem region of human immunodeficiency virus type 1 trans-activation-responsive RNA. J Virol. 1992 Mar;66(3):1688–1694. doi: 10.1128/jvi.66.3.1688-1694.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A. L., Malik S., Meisterernst M., Roeder R. G. An alternative pathway for transcription initiation involving TFII-I. Nature. 1993 Sep 23;365(6444):355–359. doi: 10.1038/365355a0. [DOI] [PubMed] [Google Scholar]
- Roy A. L., Meisterernst M., Pognonec P., Roeder R. G. Cooperative interaction of an initiator-binding transcription initiation factor and the helix-loop-helix activator USF. Nature. 1991 Nov 21;354(6350):245–248. doi: 10.1038/354245a0. [DOI] [PubMed] [Google Scholar]
- Roy S., Agy M., Hovanessian A. G., Sonenberg N., Katze M. G. The integrity of the stem structure of human immunodeficiency virus type 1 Tat-responsive sequence of RNA is required for interaction with the interferon-induced 68,000-Mr protein kinase. J Virol. 1991 Feb;65(2):632–640. doi: 10.1128/jvi.65.2.632-640.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S., Delling U., Chen C. H., Rosen C. A., Sonenberg N. A bulge structure in HIV-1 TAR RNA is required for Tat binding and Tat-mediated trans-activation. Genes Dev. 1990 Aug;4(8):1365–1373. doi: 10.1101/gad.4.8.1365. [DOI] [PubMed] [Google Scholar]
- Selby M. J., Bain E. S., Luciw P. A., Peterlin B. M. Structure, sequence, and position of the stem-loop in tar determine transcriptional elongation by tat through the HIV-1 long terminal repeat. Genes Dev. 1989 Apr;3(4):547–558. doi: 10.1101/gad.3.4.547. [DOI] [PubMed] [Google Scholar]
- SenGupta D. N., Berkhout B., Gatignol A., Zhou A. M., Silverman R. H. Direct evidence for translational regulation by leader RNA and Tat protein of human immunodeficiency virus type 1. Proc Natl Acad Sci U S A. 1990 Oct;87(19):7492–7496. doi: 10.1073/pnas.87.19.7492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheline C. T., Milocco L. H., Jones K. A. Two distinct nuclear transcription factors recognize loop and bulge residues of the HIV-1 TAR RNA hairpin. Genes Dev. 1991 Dec;5(12B):2508–2520. doi: 10.1101/gad.5.12b.2508. [DOI] [PubMed] [Google Scholar]
- Skinner M. A., Racaniello V. R., Dunn G., Cooper J., Minor P. D., Almond J. W. New model for the secondary structure of the 5' non-coding RNA of poliovirus is supported by biochemical and genetic data that also show that RNA secondary structure is important in neurovirulence. J Mol Biol. 1989 May 20;207(2):379–392. doi: 10.1016/0022-2836(89)90261-1. [DOI] [PubMed] [Google Scholar]
- Smale S. T., Schmidt M. C., Berk A. J., Baltimore D. Transcriptional activation by Sp1 as directed through TATA or initiator: specific requirement for mammalian transcription factor IID. Proc Natl Acad Sci U S A. 1990 Jun;87(12):4509–4513. doi: 10.1073/pnas.87.12.4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorge J., Cutting A. E., Erdman V. D., Gautsch J. W. Integration-specific retrovirus expression in embryonal carcinoma cells. Proc Natl Acad Sci U S A. 1984 Nov;81(21):6627–6631. doi: 10.1073/pnas.81.21.6627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taketo M., Gilboa E., Sherman M. I. Isolation of embryonal carcinoma cell lines that express integrated recombinant genes flanked by the Moloney murine leukemia virus long terminal repeat. Proc Natl Acad Sci U S A. 1985 Apr;82(8):2422–2426. doi: 10.1073/pnas.82.8.2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J. P., Pomerantz R., Bagasra O., Chowdhury M., Rappaport J., Khalili K., Amini S. TAR-independent transactivation by Tat in cells derived from the CNS: a novel mechanism of HIV-1 gene regulation. EMBO J. 1992 Sep;11(9):3395–3403. doi: 10.1002/j.1460-2075.1992.tb05418.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaishnav Y. N., Wong-Staal F. The biochemistry of AIDS. Annu Rev Biochem. 1991;60:577–630. doi: 10.1146/annurev.bi.60.070191.003045. [DOI] [PubMed] [Google Scholar]
- Weeks K. M., Ampe C., Schultz S. C., Steitz T. A., Crothers D. M. Fragments of the HIV-1 Tat protein specifically bind TAR RNA. Science. 1990 Sep 14;249(4974):1281–1285. doi: 10.1126/science.2205002. [DOI] [PubMed] [Google Scholar]
- Weeks K. M., Crothers D. M. RNA recognition by Tat-derived peptides: interaction in the major groove? Cell. 1991 Aug 9;66(3):577–588. doi: 10.1016/0092-8674(81)90020-9. [DOI] [PubMed] [Google Scholar]
- Wu F. K., Garcia J. A., Harrich D., Gaynor R. B. Purification of the human immunodeficiency virus type 1 enhancer and TAR binding proteins EBP-1 and UBP-1. EMBO J. 1988 Jul;7(7):2117–2130. doi: 10.1002/j.1460-2075.1988.tb03051.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Garcia J., Sigman D., Gaynor R. tat regulates binding of the human immunodeficiency virus trans-activating region RNA loop-binding protein TRP-185. Genes Dev. 1991 Nov;5(11):2128–2140. doi: 10.1101/gad.5.11.2128. [DOI] [PubMed] [Google Scholar]
- Zuker M. On finding all suboptimal foldings of an RNA molecule. Science. 1989 Apr 7;244(4900):48–52. doi: 10.1126/science.2468181. [DOI] [PubMed] [Google Scholar]