Figure 1.
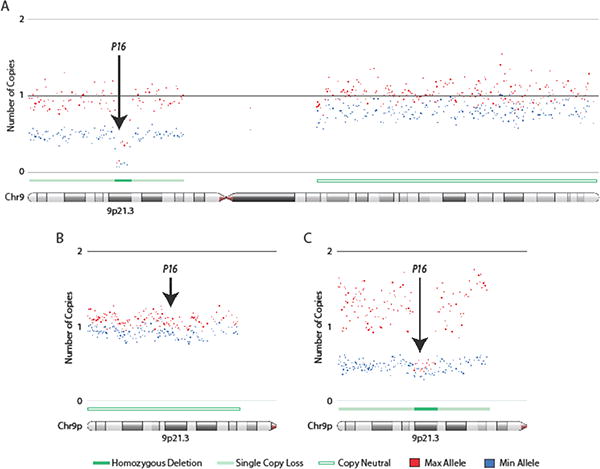
Homozygous deletions of p16 in lung adenocarcinoma tumors and cell lines. Affymetrix SNP 6 arrays were used to generate copy number profiles for lung adenocarcinoma cell lines and clinical lung tumors. Allele specific copy numbers were derived using the allele specific copy number workflow in Partek Genomics Suite (PGS) software. Copy number profiles for lung cancer cell lines were generated using a panel of 72 non-diseased HapMap individuals as a baseline while matched non-malignant lung tissue was used as a reference for defining copy number changes in tumors. Arrows indicate region of the p16 gene. (A) Focal homozygous deletion (HD) of p16 in the lung adenocarcinoma cell line, NCI-H1944. (B) Lung adenocarcinoma tumor illustrating neutral p16 copy number state. (C) Focal HD of p16 in a lung adenocarcinoma tumor. Both HDs in (A) and (C) occur in a background of single copy loss on chromosome 9p. Copy number states in regions of deletion do not drop right to zero due to cancer cell heterogeneity, aneuploidy, and/or the presence of non-malignant cells that contribute to array signals. Maximum (red) and minimum (blue) alleles are defined by copy number state. Each dot represents the smoothed copy number for 30 adjacent array probes. Copy number states (HD, single copy loss, or copy neutral) are indicated. Images were generated in PGS. Arrows indicate location of p16 gene at chromosome 9p21.3.
