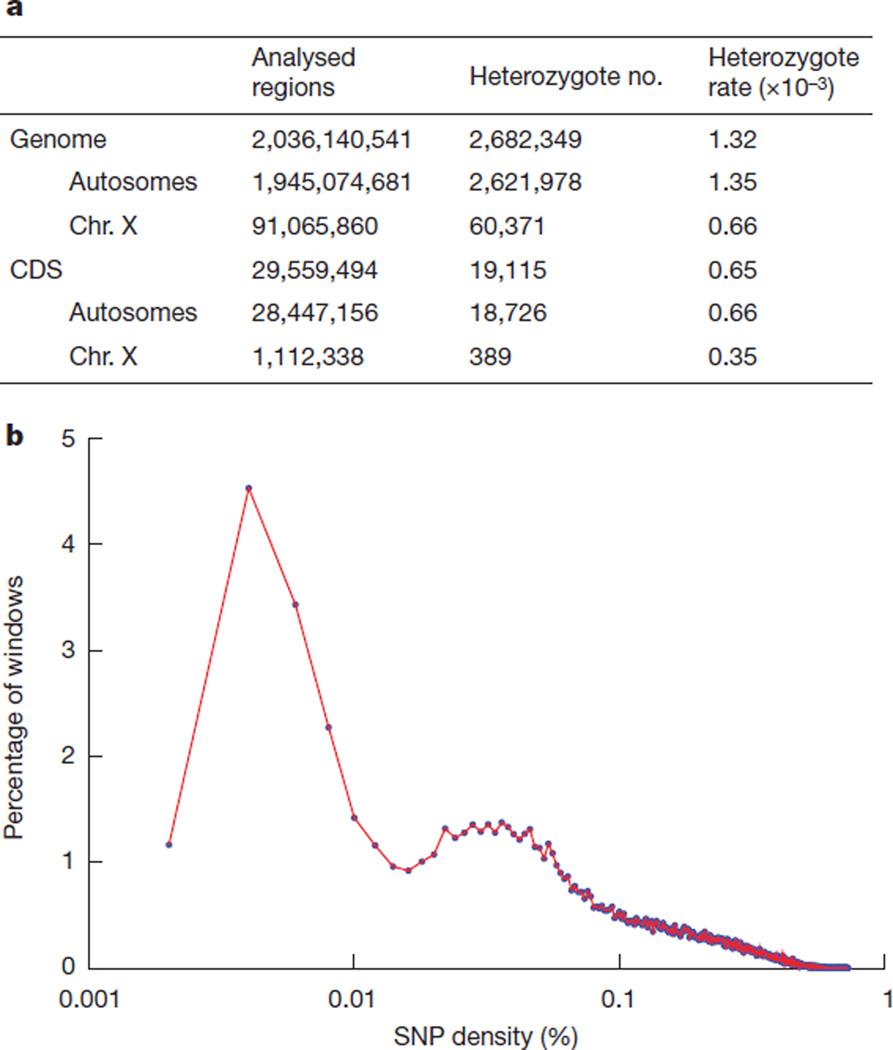
Figure 5. Panda heterozygous SNP density.
a, Statistics of identified heterozygous SNPs. Analysed regions are the genomic regions with proper unique read coverage that were used for heterozygote detection. The panda X-chromosome-derived scaffolds were identified by Blastz alignment to the dog X chromosome. b, Distribution of heterozygosity density in the panda diploid genome. Heterozygous SNPs between the two sets of chromosomes of the panda diploid genome were annotated, then non-overlapping 50-kb windows were chosen and heterozygosity density in each window was calculated.