Abstract
The vast diversity of neurons and glia of the central nervous system is generated from a small, heterogeneous population of progenitors that undergo transcriptional changes during development to sequentially specify distinct cell fates. Guided by cell intrinsic and temporal extrinsic cues, invertebrate and mammalian neural progenitors carefully regulate when and how many of each cell type is produced to form functional neural circuits. Emerging evidence indicates that neural progenitors also undergo changes in global chromatin architecture, thereby restricting the duration a particular temporal cue can act. Thus, studies of temporal identity specification and progenitor competence can provide insight into how we may use neural progenitors to more effectively generate specific cell types for brain repair.
Introduction
The complex structure of our brain – and thus its ability to perform impressive cognitive and motor functions – depends on the production of a diverse pool of neurons and glia from a relatively small number of neural progenitors during development. It is well established that spatial patterning cues can produce different types of neural progenitors, and hence different types of neurons and glia, along the rostrocaudal or dorsoventral axes of the CNS1. It is also known that individual neural progenitors give rise to distinct cell types over time, which increases neural diversity in the CNS 2. Only recently, however, has there been progress in understanding the molecular mechanisms by which individual progenitors generate a sequence of different cell types – a process that we call “temporal patterning” or “temporal identity specification” (see Box 1). An understanding of temporal patterning mechanisms is important for multiple reasons: it will illuminate how spatial and temporal cues are integrated to generate specific cell types, how aging progenitors change competence to produce different cell types over time, and may help us learn how to direct neuronal differentiation in vitro to repair the damaged or diseased brain.
Spatial identity – the aspects of progenitor identity determined by its spatial position.
Spatial identity factor/cue – a molecule that gives positional information to a cell. Example: The Drosophila engrailed gene and its orthologue En-1/En-2 in mammals help establish regional identities during development of the body plan 130.
Spatial patterning – the generation of heterogeneous neural progenitors based on their position within the developing nervous system.
Temporal patterning – the generation of distinct neural progeny in response to developmental stage-specific cues. Temporal patterning cues collectively include any intrinsic or extrinsic factor that contributes to the production of a specific progeny fate based on its birth timing (temporal identity factors, switching factors, etc).
Temporal identity – an aspect of progenitor identity determined by its birth-order.
Temporal identity factor – a factor that specifies cell fate based on the birth timing of the progeny (e.g. early or late) in multiple progenitors independent of their spatial identity. Example: The Hunchback transcription factor specifies first-born temporal identity in multiple progenitor lineages despite their different spatial identity 5.
Temporal window – the duration (either by time or number of progenitor divisions) in which a particular temporal identity factor is expressed.
Switching factor – a molecule that promotes the transition between temporal identity factors; does not directly specify cell fate. Example: Svp is required for the Hb-Kr transition in Drosophila neuroblasts 14,26.
Subtemporal factor – a factor that acts downstream of a temporal identity factor to subdivide the temporal window to multiple distinct progeny fates. Example: Nab and Sqz subdivide the last four Cas-expressing division of NB5-6 to specify distinct neurosecretory cells 20.
Cell fate determinant – a factor that specifies a particular cell fate. Such factors likely function downstream of the spatial/temporal factors.
Competence – the ability of a progenitor to generate a particular cell fate in response to a spatial or temporal identity factor. Example: Drosophila progenitors are competent to generate first-born neurons in response to Hunchback only early in their lineage 7,17.
In this review, we discuss recent advances in our understanding of temporal patterning within the Drosophila and mammalian CNS. We highlight key recent results and conserved mechanisms and discuss several important open questions. We divide temporal patterning into two processes: the specification of temporal identity (in which changing intrinsic or extrinsic cues act on a neural progenitor to specify a particular cell type) and changes in progenitor competence (through which the progenitors’ response to temporal cues and subsequently their progeny output changes).
Specification of temporal identity
We define temporal identity as the aspect of cell fate determined by its birth-order in a progenitor lineage, in contrast to the aspects of cell fate due to its position within the tissue or embryo (see Box 1). For example, “early-born” temporal identity refers to a neuronal phenotype that is generated early in the lineage rather than to a particular cell type. Spatially different neural progenitors can use the same temporal identity factor to specify distinct early- or late-born cell fates. We further define a “temporal window” as the time or the number of progenitor cell divisions during which a given temporal identity factor is expressed.
Specification of temporal identity in Drosophila
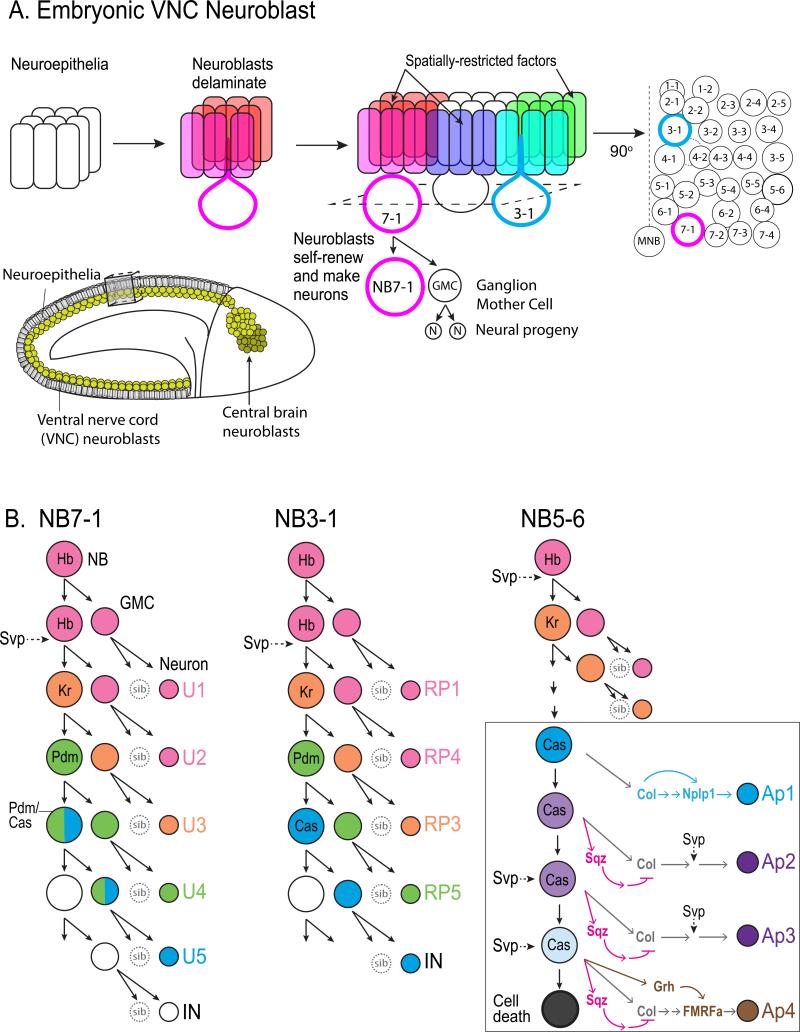
In the ventral CNS of the Drosophila embryo, 30 distinct neural progenitors, called neuroblasts, are arranged in a segmentally-repeated bilateral pattern and give rise to all neurons and glia of the nerve cord 3,4 FIGURE 1A. Neuroblasts undergo multiple rounds of asymmetric cell division. With each round, typically ~1hr per division, a smaller ganglion mother cell (GMC) ‘buds off’ and divides once more to generate a pair of neurons or glia FIGURE 1A-4. The neuroblasts form a layer at the ventral surface of the CNS, and their early-born progeny are displaced by later-born progeny, resulting in a “laminar” CNS reflecting neuronal birth order5. The major advantages of this system to study neurogenesis are that each neuroblast is uniquely identifiable by the presence of specific molecular markers and its position within a grid-like array (for example, NB7-1 always in row 7, column 1 3,4), a specific neuroblast gives rise to a reproducible set of neuronal and glial progeny, always in the same birth order6-10, and there is minimal neuronal migration6,8,9. These characteristics have allowed individual neuroblast lineages to be exceptionally well-characterized and provide a unique platform for identifying and characterizing candidate temporal identity factors.
Figure 1. Neurogenesis in Drosophila embryo ventral nerve cord neuroblast lineages.
A. (a1) Neuroblasts are specified and subsequently extruded (delaminated) from the neuroepithelia at the onset of neurogenesis. (a2) Each individual neuroblast can be uniquely identified based on its stereotyped position within the nerve cord and its expression of spatially restricted factors. (a3) Neuroblasts are named according to their row and column within the neuroblast “grid.” Thus, NB7-1 (red) is present in the seventh row and occupies the first column position, whereas NB3-1 (blue) is positioned in the third row. (a4) Each neuroblast undergoes a series of asymmetric divisions that give rise to a self-renewed neuroblast and a differentiating ganglion mother cell (GMC). The GMC divides again to generate two neural progeny. The inset shows a Drosophila embryo at stage 9, at the onset of neuroblast delamination. Neuroblasts are shown in yellow along the ventral nerve cord just beneath epithelial layer. b-d. The lineages of NB7-1, NB3-1, and NB5-6 neuroblasts are shown. Each sequentially expresses the temporal identity factors Hunchback (Hb), Kruppel (Kr), Pou domain protein (Pdm), and Castor (Cas) and give rise to a unique lineage of neural progeny in a stereotyped birth order. The COUP-family nuclear receptor, Seven-up (Svp) is transiently expressed in neuroblasts to regulate timing of temporal fate determinant expression. D, inset. At the end of the NB5-6 lineage, Castor initiates a subtemporal transcriptional cascade by activating Squeeze (Sqz, red rectangle) and Grainyhead (Grh, green rectangle). The delay in the accumulation of Sqz and Grh as NB5-6 divides results in distinct neuronal subtypes generated during the Castor window. At the end of the lineage, NB5-6 undergoes cell death.
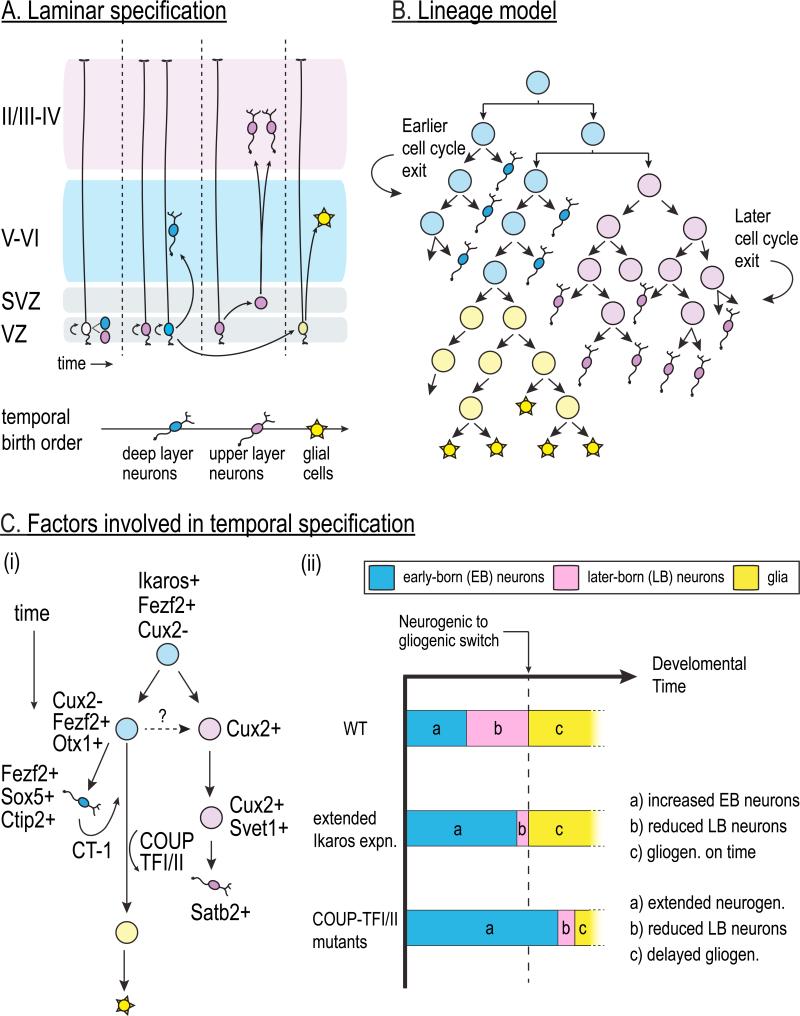
Figure 4. Temporal fate specification in mammalian cortex.
A. The six layers of the mammalian cortex are generated by the sequential production of distinct types of neurons that migrate to progressively more superficial layers in an “inside-out” fashion. Deep-layer neurons (blue) are born first from the ventricular zone (VZ) radial glia. Subsequently, upper-layer neurons (red) are born from a subset of VZ radial glia as well as the intermediate progenitors in the subventricular zone (SVZ) that are born from the VZ progenitors. Finally, glia (green) are born after the neurogenic period ends.
B. Progenitors that give rise to deep layer neurons (blue) exit the cell cycle earlier than the progenitors that primarily give rise to the upper layer neurons (red).
C. Factors that function in laminar cell fate are shown on the left. A multipotent progenitor gives rise to more restricted lineages that preferentially generate deep (Cux2-negative) or superficial (Cux2-expressing) cortical neurons. COUP-TFI and II as well as extrinsic signals like CT-1 act on progenitors to switch from neurogenesis to gliogenesis. It is currently unknown whether all Cux2-expressing SVZ progenitors that generate upper layer neurons are derived from the Cux2-expressing VZ progenitors, or whether these are separate progenitor pools. Extension of Ikaros expression in cortical progenitors or loss of COUP-TF results in an expansion of early-born cortical phenotypes (a, blue) at the expense of later-born phenotypes (b, red). However, extension of Ikaros affects the balance of neuronal fates, but does not affect timing of gliogenesis (c, green).
The first candidate temporal identity factors came from the observation of laminar expression of the transcription factors Hunchback (Hb), Pou domain protein 1 and 2 (Pdm; Nubbin and Pdm2, Flybase), and Castor (Cas), which are expressed in deeper and more superficial neuronal layers, respectively in the mature CNS 11. Subsequently, it was shown that most neuroblasts sequentially express Hb, the transcription factor Kruppel (Kr), Pdm, and Cas as they undergo multiple rounds of cell division 5. Thus, sequential expression of different transcription factors in neuroblasts leads directly to the laminar expression observed in the mature CNS.
The embryonic neuroblast 7-1 (NB7-1) has the best characterized lineage of all Drosophila neuroblasts, producing five distinct types of motoneurons (U1-U5) during its first five divisions FIGURE 1B. Hb is an Ikaros family zinc-finger transcription factor that is necessary and sufficient to specify earliest-born neuronal identity in multiple neuroblast lineages: NB7-1, NB7-3, and NB3-1 5,7,10,12-15. Although the specific characteristics of the first-born progeny differ between neuroblast lineages, cells specified with an early-born identity use a discrete neuronal enhancer 16 to maintain active transcription of the hb gene as a molecular marker of their early temporal birth 17,18. Next, another zinc-finger transcription factor, Kr, specifies the second temporal fate in multiple neuroblast lineages 5,12,13 FIGURE 1B-C. Both Hb and Kr expression are maintained in neuronal progeny, but what role Hb and Kr play in postmitotic neurons is not known.
The roles of the later candidate temporal identity factors Pdm and Cas have been characterized in multiple neuroblast lineages, with different results in each lineage tested. In the NB7-1 lineage FIGURE 1B, Pdm is necessary and sufficient to specify the U4 motoneuron fate, and Pdm and Castor together specify the U5 motoneuron identity 19. In the absence of cas, U5 neurons are lost, whereas overexpression of Pdm and Cas together generate extra U5 neurons 19. In the NB3-1 lineage, however, Pdm has no detectable role in specifying the RP5 neuron born during the Pdm phase of expression. Instead, Pdm is required to repress Kr: in its absence, Kr expression is extended, resulting in the production of extra Kr-expressing RP3 motoneurons FIGURE 1C10. Thus, Pdm acts to specify cell fate in NB7-1 and as a “switching factor” in NB3-1 to regulate the timing of Kr expression. In both NB7-1 and NB3-1 lineages, Cas is required to close the temporal window that specifies late-born neurons, and loss of cas results in extra late-born neuron cell types 5,10, indicating that Cas can act as a switching factor in addition to its fate-specifying functions.
The role of Cas, has perhaps been the best characterized in the lineage of NB5-6, in which the Apterous transcription factor is expressed in the last four neurons in the lineage 20 FIGURE 1D. Cas is expressed by NB5-6 in a broad temporal window that spans ten divisions and includes the last four divisions of the lineage, during which it activates the transcription factor Collier and specifies Apterous-expressing neurosecretory neurons. Cas initiates a feed-forward transcriptional pathway by activating downstream factors such as Squeeze that help to establish the individual identity of the Apterous-expressing neurons 20. Thus, neuroblast temporal windows can be established by one set of factors and be further subdivided by “subtemporal factors” through feedforward and feedback transcriptional regulation that increases neural diversity 20. The transcription factor Grainyhead (Grh), is a candidate temporal identity factor following Cas in multiple neuroblasts 21-23. In the embryonic NB5-6 lineage, Grh is required to specify the last-born, FMRFa-expressing neurosecretory neuron 20, and Grh also plays a role in specifying temporal fate in intermediate progenitors of the larval Type II NB 24(see below). More evidence to compare the role of Grh in specifying cell fate 20,24 separate from its role in cell cycle 21-23 would be needed to determine its function as a temporal identity factor.
Collectively, the accumulating evidence indicates that Hb, Kr, and Cas are bona fide temporal identity factors, as they specify the identity of neuroblast progeny based on their birth timing in multiple neuroblast lineages. Analysis of more neuroblast lineages is required before determining whether Pdm is also a temporal identity factor. In addition to fate-specification, both Pdm and Castor appear to function as switching factors depending on the lineage, perhaps in response to activity of lineage-specific spatial patterning cues.
What regulates the timing of temporal identity factor expression? Misexpression experiments show that each temporal identity factor can activate the next factor in the pathway and repress the factor that follows5,21, but loss of Hb or Kr does not affect the production of the later-born cells 5, suggesting an independent mechanism contributes to the sequential expression of the temporal identity factors. The Hb-Kr transition requires both neuroblast cytokinesis and the expression of COUP-family Seven-up (Svp) orphan nuclear receptor 14,25,26. Svp is expressed in two temporal waves in embryonic neuroblast lineages 25 and appears to regulate the timing of multiple events, consistent with a role as a switching factor FIGURE 1B-D. Its initial wave represses hb transcription to allow the Hb-to-Kr transition, and recent work shows that its re-expression at a later stage in the NB5-6 lineage subdivides the broad, ten-division Cas expression window into multiple distinct neural fates 25,27. Dissociated embryonic neuroblasts still sequentially express temporal identity factors as they would in vivo 18,21, and remarkably, the Kr–Pdm–Cas transitions can occur in cell cycle-arrested neuroblasts 18, indicating a robust, neuroblast-intrinsic timing mechanism that is independent of cell cycle progression 18,21. The molecular nature of the Kr-Pdm-Cas timer mechanism remains unknown.
Larval type I neuroblasts have lineages similar to embryonic neuroblasts, and also undergo temporal transitions that expand neural diversity, but use different transcription factor cascades. Here we discuss three examples: the AD neuroblast, the four mushroom body neuroblasts, and the optic lobe medulla neuroblasts. The AD neuroblast generates a different projection neuron with each cell division; Kr is expressed for just one cell division to specify VA7l neuronal identity, but there is no known role for Hb, Pdm, or Cas 28. The four mushroom body (MB) neuroblasts, and many other larval neuroblasts, generate multiple neurons expressing the transcription factor Chinmo, followed by a series of smaller neurons expressing Broad-Complex transcription factors 23,29 FIGURE 2A-B. Chinmo protein levels within the postmitotic neurons decline over time in a graded manner. In mushroom body neuroblast lineages, high Chinmo specifies early-born γ neuron identity, while low Chinmo specifies mid-born α’β’ neuronal identity. The Chinmo protein temporal gradient is generated post-transcriptionally through the expression of the let-7 and mir-125 microRNAs in the late-born neurons 30. Intriguingly, Svp appears to reprise its embryonic role as a switching factor in larval neuroblasts: larval neuroblasts transiently express Svp just prior to the switch from the production of Chinmo-expressing neurons to the production of Broad-Complex-expressing neurons, and Svp mutant neuroblasts never make this switch 23. Lastly, recent exciting work has revealed a completely novel cascade of temporal identity factors in the optic lobe medulla neuroblasts. These neuroblasts give rise to a diverse array of neurons in the visual processing regions 31-33. Two elegant genetic studies recently showed that most optic lobe neuroblasts sequentially express the transcription factors Homothorax, Eyeless, Sloppy-paired, Dichaete and Tailless and contribute to ~40,000 neurons of more than 70 distinct subtypes in the medulla 34,35. These studies give us a glimpse of the complexity with which temporal fate may be regulated in a context-dependent manner.
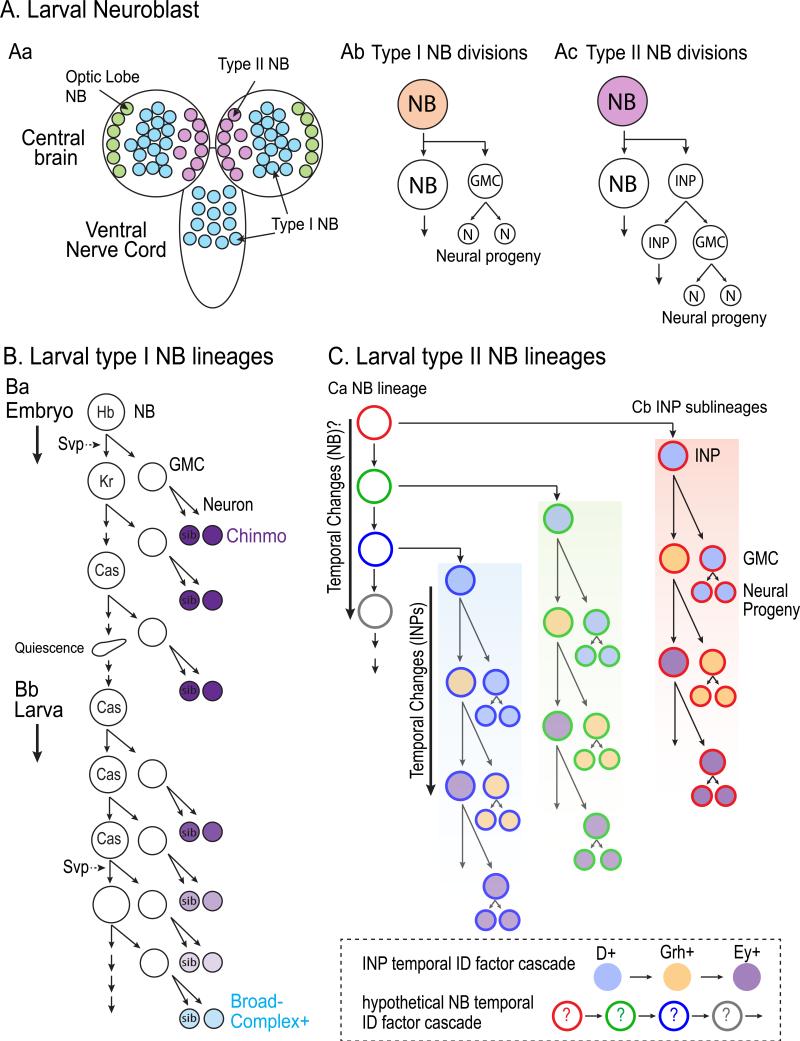
Figure 2. Neurogenesis in Drosophila larval central brain neuroblast lineages.
A. (Aa) The larval central brain harbors roughly 100 neuroblasts in addition to the optic lobe neuroblasts (green). These are divided into type I (blue) and type II (red) neuroblasts. (Ab) Type I neuroblasts of the central brain and ventral nerve cord (blue) divide similarly to embryonic ventral nerve cord neuroblasts shown in Figure 1 by generating a ganglion mother cell (GMC) that divides to produce two neural progeny. (Ac) Type II neuroblasts divide to give rise to an intermediate progenitor (INP) that undergoes additional “neuroblast-like” asymmetric divisions, thus greatly amplifying the number of neural progeny.
B. Type I neuroblast lineages at the larval stages express Cas and typically give rise to a series of Chinmo-expressing neurons followed by a series of Broad-Complex-expressing neurons. Chinmo expression is downregulated in the postmitotic progeny in a gradient, with early-born neurons expressing the highest Chinmo levels. (Ba) While Svp regulates the Hb-to-Kr transition in the embryo, (Bb) it is re-expressed in larval neuroblasts to regulate the temporal transition from Chinmo expression to Broad-Complex expression.
C. INPs generated from a Type II neuroblast sequentially express Dichaete (D, blue), Grainyhead (Grh, orange), and Eyeless (Ey, purple) to temporally specify distinct neural progeny. Given that neural progeny born early in the neuroblast lineage are different from those born later, it is likely that the neuroblast itself undergoes temporal transitions that are inherited by the INPs (hypothetical NB temporal identity factors are depicted by colored circle outlines).
Larval type II neuroblasts have a more complex lineage: they sequentially produce intermediate neural progenitors (INPs) that themselves undergo multiple molecularly-asymmetric self-renewing divisions to produce a series of ~ 6 GMCs that terminally divide to generate two neurons or two glia 36-38 (FIGURE 2Abc-Ac). Thus, compared to type I neuroblasts which produce GMCs directly and generate ~100 neurons per lineage, type II neuroblast lineages contain an amplifying progenitor population that greatly their neural output to ~ 600 neurons of over 60 subtypes 24,36-41. Where does this neural diversity originate within the type II neuroblast lineages? It has recently been shown that nearly all INPs transition through a three transcription factor cascade as they divide – Dichaete, Grainyhead, and Eyeless – and these factors specify early-to-late temporal identity within multiple INP sub-lineages 24 (FIGURE 2Ci). In addition to the temporal transitions within INPs, distinct neurons and glia are also produced as type II neuroblasts age over time 24,42, suggesting that currently unknown temporal identity cascades likely exist within the parental neuroblast as well. Taken together, it is likely that the combinatorial activity of two independent temporal cascades – one within the neuroblast and one within the INPs – is used to generate neural diversity of the adult brain central complex. How these two temporal axes intersect, and how they may be further regulated by spatial patterning cues are fascinating questions for future work.
The parallels between the Drosophila type II neuroblast lineages and mammalian neural stem cell lineages are intriguing. For example, neural stem cells of the adult subventricular zone, the largest germinal zone of the adult mammalian brain, generate INP-like transit-amplifying progenitors before generating diverse subtypes of olfactory bulb neurons 42. Additionally, recent work identified a second population of asymmetrically-dividing and self-renewing progenitors, similar to the ventricular zone radial glia, in the outer subventricular zone (OSVZ) of the developing human brain that may have greatly expanded cortical size and complexity during evolution 43,44. Whether mammalian progenitor or INP lineages undergo temporal identity transitions, and how they contribute to the generation of neural diversity, is an important area for future investigation.
These studies pave the way for several research avenues. Perhaps the two most important will be to investigate how spatial cues and multiple temporal cues are integrated to generate neural diversity, and how temporal identity factors govern neuronal terminal differentiation. An attractive possibility is that spatial and/or temporal patterning factors in the neuroblast initially establish a chromatin state that determines which target genes becomes transcriptionally active in response to downstream temporal patterning factors, and such chromatin states can subsequently be inherited by the postmitotic progeny.
Specification of temporal identity in mammals
Neural progenitors in the developing mammalian CNS also generate distinct neural progeny in a stereotyped birth order. Here we focus on three examples: the ordered production of retinal cell types, cortical laminar identities, and the switch from neurogenesis to gliogenesis. The sequential production of visceral motoneurons and serotonergic hindbrain neurons has been reviewed elsewhere 2,45,46.
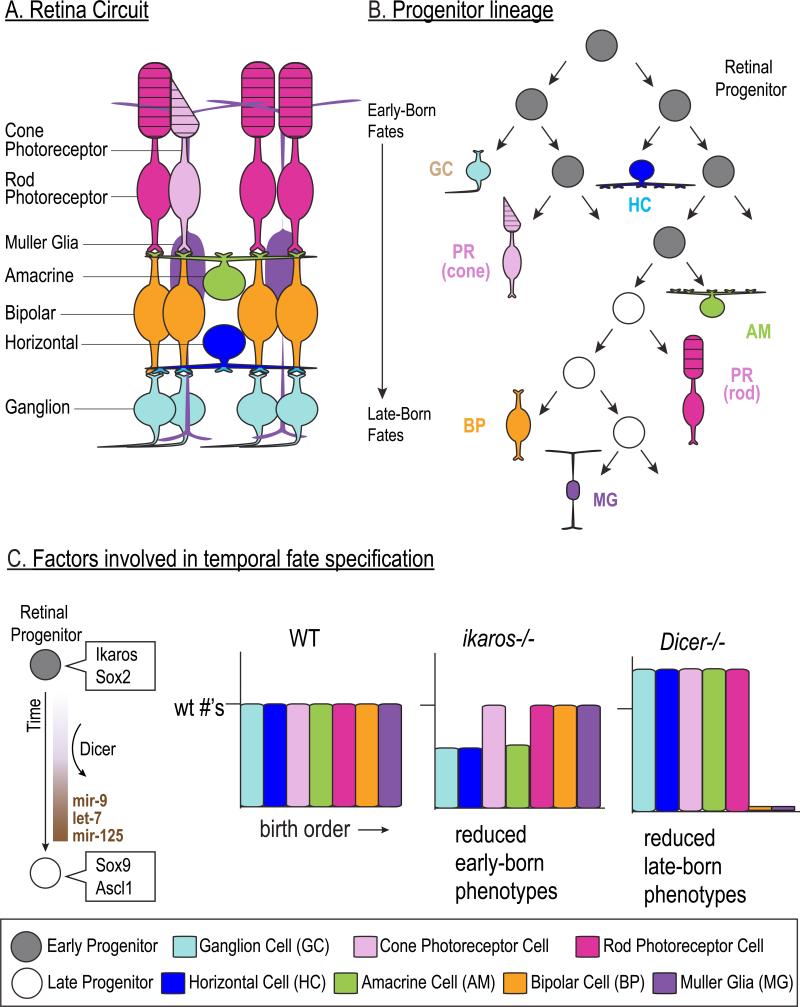
In vivo lineage tracing has shown that individual neural progenitors in the vertebrate retina are multipotent and give rise to distinct cell types in a characteristic birth order 46-49 FIGURE 3. Transcriptome analysis of single retinal progenitor cells from different developmental stages revealed the sequential expression of transcription factors related to the fly temporal identity factors Hb, Kr, Pdm, and Cas 50. The zinc finger transcription factor, Ikaros, is expressed by young retinal progenitors and is required to specify the early-born cell types 51 FIGURE 3C, remarkably similar to the role of its fly orthologue, Hb 5. Despite the ordered production of distinct cell types during retinal development, cell lineage-tracing studies of individual progenitors show considerable variability in the number and the composition of neural progeny. Clonally-cultured retinal progenitors appear to follow stochastic patterns to decide between self-renewal and differentiation, and the order of retinal cell types produced within each individual progenitor lineage does not necessarily follow the order observed across the whole progenitor population 52. Retinal progenitors from earlier developmental stages were biased to undergo more self-renewing divisions than those from later stages, however, indicating a cell-intrinsic shift in division mode over time (52, reviewed in 53). Alternatively, it is possible that there are multiple different progenitor subtypes that each has a different, but highly reproducible cell lineage (similar to fly neuroblasts). Consistent with this idea of progenitor heterogeneity is the recent finding that Cadherin-6 and Olig2 mark subsets of retinal progenitors that bias the fate of their neuronal progeny 54,55. How spatial, temporal, and stochastic mechanisms might integrate to balance diversity and order is a fascinating question.
Figure 3. Temporal fate specification in mammalian retina.
A. Schematic illustration of the main cells types in the retina and their organization within the retinal circuit. The retina is comprised of six major classes of neurons and one type of glia (the Muller Glia).
B. Retinal progenitors give rise to these distinct cell types in an overlapping but sequential order. Ganglion cells are generated first by early retinal progenitors (light blue) and bipolar cells (orange) and Muller glia (purples) are born last from late progenitors.
C. Several molecular factors are expressed in either early (gray) or late (white) progenitors and can determine the temporal phenotypes of the progeny. Dicer is required for the expression of several microRNAs that regulate the temporal transition of the progenitors to produce late-born cell fates. In the mouse retina lacking the transcription factor Ikaros, there is a decrease in the number of cells with early-born fates, although the cone photoreceptors (pink bar) are not affected. In contrast, in mouse retina lacking Dicer, a key enzyme involved in microRNA processing, there is a loss of the late-born cell fates.
The mammalian cerebral cortex provides a most striking example in which radial migration of neuronal progeny based on birth order dictates cortical laminar organization 56,57. Ventricular zone (VZ) progenitors in the pseudostratified neuroepithelium initially divide symmetrically to expand the progenitor pool. Elegant time-lapse microscopy studies revealed that radial glia, the progenitors of the VZ, then directly give rise to neurons while undergoing self-renewing divisions, and subsequently give rise to intermediate progenitors that undergo symmetric neurogenic divisions within the subventricular zone (SVZ) to produce two more progenitors or two neurons 58,59. Postmitotic neurons born from progenitors in the VZ and the SVZ radially migrate to the cortical plate, with later-born neurons climbing past the earlier-born neurons, resulting in six distinct layers formed in an inside-out fashion based on birth order (reviewed in more detail in 56,57) FIGURE 4A-B.
Although both the VZ and SVZ generate cortical neurons, studies of genes expressed in the VZ (e.g. Pax6 and Otx1) and SVZ (e.g. Cux1/2 and Svet1) – and mice mutant for those genes – have led to the model that VZ progenitors generate deep layer VI-V neurons, and SVZ progenitors generate superficial layer IV-II neurons 60-63. Cux2, however, is detected in a small subset of progenitors in the VZ as early as E10.5, prior to the formation of the SVZ, which led to the suggestion that Cux2-expressing progenitors in the VZ may be from the outset committed to generating the SVZ and later-born upper layer neurons 63. Consistent with this model, in utero fate-mapping shows that Cux2-expressing VZ progenitors are fate-restricted to give rise to upper cortical layer neurons, regardless of niche or birthdate, and that superficial versus deep laminar fate is the result of timed cell cycle exit of progenitors rather than sequential specification 64. What is not yet clear is whether Cux2-negative VZ progenitors can produce deep layer neurons before beginning to express Cux2 FIGURE 4C. Other lineage-tracing studies have, however, provided evidence for individual progenitors that give rise to neurons of both upper and lower layers. For example, in utero electroporation of green fluorescent protein (GFP)-encoding retroviruses to label progeny of single VZ radial glia has shown that ontogenetic radial clones of excitatory neurons span deep and superficial cortical layers 65. These sibling neurons form synaptic connections that foreshadow mature cortical circuitry, suggesting that a single VZ progenitor can give rise to neurons in both upper and lower layers and can contribute to the columnar microcircuit.
It is clearly important to understand how cell cycle regulation intersects with temporal generation of neuronal subtypes. Previous studies have shown that loss of Cux2 in the cortex results in increased proliferation of SVZ progenitors and an overproduction of upper-layer cortical neurons; overexpression of Cux2, on the other hand, promotes cell autonomous cell cycle exit of neural progenitors in vitro 66. These observations appear to be at odds with the finding that Cux2-expressing progenitors remain mitotically active for longer than those lacking Cux2, and that these are the progenitors that generate upper-layer neurons 64. It is possible that Cux2 may function differently in VZ and SVZ progenitors or may change its function over time. This appears to be the case for the zinc-finger transcription factor, Sal1, which is highly expressed in cortical progenitors but downregulated in differentiating neurons 67. In Sal1 knockout mice, progenitors in the early stages of corticogenesis, predominantly the VZ radial glial cells, prematurely exit the cell cycle and differentiate into neurons, whereas progenitors at later stages, predominantly the SVZ intermediate progenitors, re-enter the cell cycle without differentiating, resulting in fewer upper-layer neurons 67. Another regulator of progenitor cell cycle is Gde2, a six-transmembrane protein that is detected throughout corticogenesis in postmitotic neurons 68. In Gde2 mutants, progenitors fail to exit the cell cycle until the end of the normal neurogenic period, and differentiate en masse into upper neuron identities at the expense of early-born identities 68. Thus, Gde2 is an extrinsic regulator of progenitor cell cycle exit (through feedback from the neuronal progeny) and temporal identity switching. While the role of Sonic hedgehog (Shh) in spatial patterning is well established, increasing evidence implicates Shh in the regulation of the cell cycle and temporal identity. In the Xenopus retina, Shh regulates the length of the progenitor cell cycle, which in turn regulates expression of several microRNAs important in specifying temporal cell fate 69. In the chick spinal cord, Shh promotes progenitor pool expansion of the at the expense of neuronal differentiation, and thus, the timing of motoneuron formation 70. Shh, SalI, and Gde2 are important demonstrations of how extrinsic and intrinsic cues can regulate the timing of progenitor differentiation.
In recent years tremendous progress has been made in identifying factors that are expressed in a cortical layer-specific manner and determining how they specify laminar fates (FIGURE 4C). Fezf2, SOX5, and Ctip2 are transcription factors required to specify the early-born projection neurons that occupy the deep cortical layers and project to subcortical regions 71-75. Fezf2 promotes specification of deep layer subcortical projection neurons in part by repressing the chromatin remodeling protein Satb2 74. Conversely, Satb2 promotes superficial layer, callosal projection neuron identity by repressing Ctip2 expression 71,76. The POU-domain transcription factors Brn1 and Brn2 also play an important role in generating upper-layer neurons 77. Recent important work using combinatorial deletion of these laminar fate-specifying factors uncovered complex, cross-inhibitory genetic interactions between cell fate determinants in the postmitotic neurons to actively repress alternate fates and execute the developmental stage-appropriate transcriptional program 78. Interestingly, Satb2 and Fezf2 regulate genes implicated in neurological disorders, providing an entry point to studying complex diseases 78. Whereas Sox5 and Ctip2 act in postmitotic neurons to promote deep layer-specific phenotypes, Fezf2 is transiently expressed by young VZ progenitors and is maintained in the early-born deep layer V/VI neurons 75 much like the Hb and Kr temporal identity factors in fly neuroblasts 5. Although Fezf2 is necessary and sufficient to induce early cortical neuron phenotypes 74,75, whether it functions as a bona fide temporal identity factor or as a cell fate determinant for a specific cortical neuron subtype will depend on whether it functions to specify early-born phenotypes in multiple neural stem cell lineages in the cortex or elsewhere. Ikaros, in contrast, appears to fulfill the criteria to be a temporal identity factor, because in addition to its role in specify early temporal fate in the retina 51, recent work shows that it can also induce early-born neuronal fate in the cortex 79. When Ikaros expression is genetically maintained in progenitors, there is a sustained increase in the generation of early-born, deep layer cortical neurons and a decrease in the upper layer neurons 79 FIGURE 4C. How Fezf2 and Ikaros work together to specify early-born temporal identity remains to be determined. In addition to those mentioned above, numerous other transcription factors show cortical layer-specific expression patterns 57, and it would be important to understand how these factors fit into the transcriptional network both in the progenitor and the postmitotic neurons to establish sharp boundaries of temporal identities.
One transition in cell fate observed in multiple regions of the developing CNS is the switch from neurogenesis to gliogenesis 80,81. Previous studies have found that the neurogenic factor Neurogenin 2 can inhibit gliogenesis 82; conversely, the gliogenic factor Sox9 is required to end neurogenesis and promote gliogenesis 83. Extrinsic mechanisms also play an important role: signaling by the cytokines ciliary neurotrophic factor (CNTF), leukemia inhibitory factor (LIF), and cardiotrophin-1 (CT-1) induce astrogenesis by activating the glial genes glial fibrillary acidic protein (GFAP) and s100β 84-87. CT-1 is secreted by newly born cortical neurons, indicating that the onset of gliogenesis involves feedback regulation 84 FIGURE 4C. Other signaling pathways, such as Notch and BMP signaling, also promote gliogenesis (reviewed in 88). These results suggest that neurogenic and gliogenic cell fate programs are closely interconnected via multiple cell intrinsic and extrinsic mechanisms. Intriguingly, recent work has shown that the Coup-TFI and Coup-TFII nuclear receptors, orthologues of Drosophila Svp, function as a “timer” that switches progenitors from neurogenesis to gliogenesis. They are transiently expressed in neural progenitors near the end of the neurogenic phase, and their loss prolongs neurogenesis at the expense of gliogenesis 89. Additionally, COUP-TFI has also been implicated in switching from early-born to late-born cortical neurons 90. Thus, Svp and COUP-TFI and II appear to have a conserved role as switching factors in both Drosophila and mammalian neural progenitors FIGURE 4C.
MicroRNAs have recently been added to the repertoire of factors that contribute to temporal fate specification FIGURE 3C. Conditional deletion of Dicer, a key microRNA-processing enzyme, in progenitors results in an increase in early-born neuronal phenotype and a decrease in late-born phenotypes in both cortex and retina 91-94 (recently reviewed in reference 95), and a loss of late-born glia in the spinal cord while leaving early-born motoneurons intact 96. Ikaros, like its Drosophila orthologue Hb, promotes early-born cell fate in both the cortex and the retina. In the retina, Ikaros mRNA is expressed throughout development, whereas its protein is detected only in the early progenitors 51, suggesting posttranscriptional regulation, perhaps through microRNA function. In C. elegans, in which microRNAs have a well-documented role in regulating the timing of developmental transitions 97, the let-7 family of microRNAs must degrade hunchback-like (hbl) RNA, the C. elegans orthologue of Drosophila hb, to allow the transition from the L2 larval stage to L3 98. Whether Ikaros and Dicer-mediated regulation of temporal identity are part of the same or parallel pathways, and whether other members of the Ikaros family function in temporal fate specification are still unknown. Recent work has further shown that microRNAs function at multiple stages of neuronal differentiation. When Dicer is deleted in postmitotic neurons of the cortex, there is a reduction in dendritic branching, but the laminar organization is normal 99. In Drosophila the let-7 and mir-125 microRNAs are expressed in some larval neuroblast lineages by late-born neurons and are required to specify late neuronal temporal identities by regulating the levels of Chinmo protein expression 30. Thus, microRNAs appear to play an important role in temporal fate specification in both the progenitor and the postmitotic progeny, and in multiple organisms.
Changes in progenitor competence
In addition to the transcriptional changes that neural progenitors undergo to specify distinct temporal cell fates, the competence of neural progenitors to specify particular fates also changes during development. While gradually losing the ability to specify earlier-born cell fates (competence restriction), neural progenitors acquire competence to make later-born cell types. Thus, a given neural cell type can be specified during a very limited time window. Investigating how competence is regulated is critical to our understanding of brain development and ongoing efforts to generate specific neural cell types from induced pluripotent stem cells.
Changing progenitor competence in Drosophila
The best characterized model for studying progenitor competence is the embryonic NB7-1, for which there are markers for each of the neuronal progeny (U1-U5) specified by temporal identity factors 3,5,7,12 FIGURE 1B. Pioneering work showed that although Hb normally specifies U1/U2 fates during the first two neuroblast divisions 6,8,9, pulses of ectopic Hb expression in NB7-1 at progressively later stages can induce extra U1/U2 neuron generation until its fifth division 7 FIGURE 5A. Similarly, competence to respond to Kr and specify the U3 fate is also lost after the fifth division, showing that NB7-1 has an early competence window to respond to both Hb and Kr 12,13.
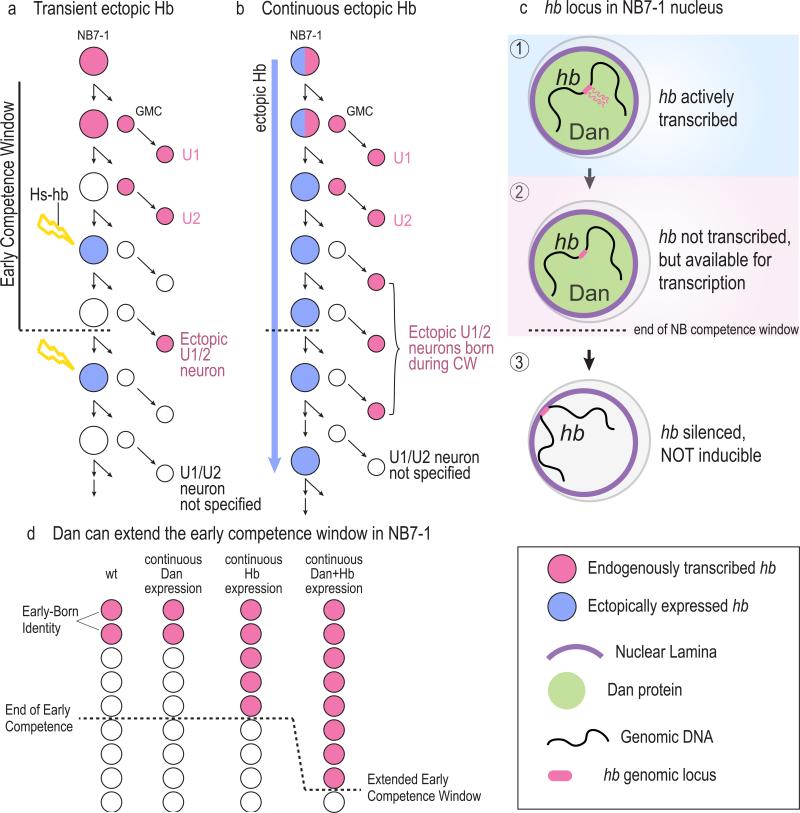
Figure 5. Reorganization of the neuroblast genome regulates competence transition in Drosophila embryos.
A. The Hb temporal identity factor is expressed in NB7-1 of the fly embryonic nerve cord for the first two divisions (red), giving rise to the U1/U2 early born motoneurons, which maintain Hb expression. A transient pulse of ectopic Hb (blue) can induce the neuroblast to produce an ectopic U1/U2 neuron up to the fifth division (a). These first five divisions are called the “early competence window.” After this window ends (dotted line), NB7-1 is no longer competent to respond to ectopic Hb and cannot specify early-born neuronal fate.
B. If ectopic Hb (blue) is continuously expressed in the neuroblast, only the postmitotic progeny born during the early competence window, and not those born after, will activate endogenous hb transcription (red).
C. (c1) During the first two divisions when hb is actively transcribed, the hb genomic locus is positioned in the nuclear interior. (c2) During the subsequent three divisions, the hb gene is transcriptionally inactive, but is still positioned in the nuclear interior and is amenable for activation in the progeny. (c3) At the end of the five division competence window, the hb locus becomes repositioned to the nuclear lamina where it is permanently silenced and is no longer inducible. The nuclear factor Dan (green) is expressed in the neuroblast during the early competence window (c1 and c2), and its downregulation (c3) is required for hb gene repositioning to the lamina.
D. Dan can extend the NB7-1 early competence window. Continuous expression of Hb alone results in the specification of early-born identity only during the early competence window. Continuous expression of Hb and Dan together results in prolonged NB7-1 competence to specify early-born identity.
What closes the early competence window? It was initially thought that sequential expression of the temporal identity factor genes restricts neuroblast competence, as early work suggested that continuous expression of Hb can prevent Cas expression and extend the competence window 7. However recent work from our lab has revised this conclusion 17. We have found that Hb cannot extend neuroblast competence, and that temporal fate specification and competence are regulated independently FIGURE 5B. We have shown that neuroblasts lose competence to specify early-born fate by undergoing a developmentally-regulated reorganization of the genome that repositions the hb gene to the nuclear lamina 17, a gene-silencing hub 100-102 FIGURE 5C. As described above, NB7-1 normally specifies hb-transcribing, early-born identity neurons for the first two divisions, and yet remains competent to specify early-born fate for an additional three divisions. This suggested that the hb gene locus, though transcriptionally turned off at the second neuroblast division, undergoes a subsequent transition to a permanently silenced state at the fifth division FIGURE 5B-C. It is well known that genes occupy non-random subnuclear positions that can impact their transcriptional states 103. By employing in vivo DNA FISH (fluorescent in situ hybridization), we discovered that the repositioning of the hb locus to the nuclear lamina coincided with the end of the competence window at the fifth division, three cell divisions after the end of hb transcription 17 FIGURE 5C. Genetic disruption of the nuclear lamina reduced hb gene-lamina association and increased the probability of NB7-1 producing an extra hb-transcribing neuron, indicating that the nuclear lamina is essential for permanently silencing the hb gene 17.
How is neuroblast competence regulated? Previous work had identified the partially redundant Distal antenna (Dan) and Dan-related (Danr) proteins, members of the CENP-B/transposase family of proteins 104, as regulators of embryonic neuroblast temporal identity 25. Their expression in neuroblasts is transient and is rapidly downregulated coincident with hb gene movement to the lamina and the end of the early competence window 17 FIGURE 5C. While prolonged expression of Dan alone in neuroblasts has no effect on the timing of hb transcription or number of early-born neurons, the hb genomic locus fails to move to the nuclear periphery, and the early competence window to specify the early-born identity is extended FIGURE 5D. The extension in competence is revealed only when expression of the temporal identity factor Hb is also prolonged in the neuroblast together with Dan. This shows that progenitor competence and temporal identity are regulated by two distinct mechanisms: Dan regulates competence and Hb specifies temporal identity. Interestingly, Dan is expressed in neuroblasts in two phases: first in newborn neuroblasts, and then again in the late embryonic neuroblasts 25. The temporal identity factor Kr is also expressed twice in the NB7-1 lineage, once during the first Dan window (where it specifies U3 motor neuron) and again during the second Dan window when interneurons are being generated 12. An attractive model is that Dan is used to generate two competence windows, allowing the same temporal identity factor to act on a distinct genome architecture or epigenetic landscape to produce different outcomes in each competence window. Further experiments are needed to determine the molecular mechanisms of Dan function, its target genes during its early and late phase of expression, and how its expression is regulated. The near synchrony with which Dan is downregulated in neuroblasts 17, which delaminate at different times and vary widely in the lineages lengths 5,105, suggest that a global extrinsic signal may play a role in regulating Dan expression and neuroblast competence.
Neuroblast competence is also regulated by the Polycomb Repressive Complexes (PRC), which promote heritable gene silencing during development 106. Overexpressing Kr in a genetic background of reduced Polyhomeotic or Suppressor of zeste 12, components of PRC1 or PRC2, respectively, led to greater numbers of ectopic Kr-specified fates compared to overexpressing Kr alone 13. Conversely, overexpressing Polyhomeotic suppressed this effect. Determining the relationship between Dan- and PRC-regulated competence will give further insight into the mechanism of progenitor competence.
Recent evidence from C. elegans suggests that changes in genome architecture may be a common characteristic of competence transitions. It was recently shown that decompaction of chromatin at the lys-6 miRNA locus is required to make only one of two bilateral neurons competent to transcribe lys-6 at a later developmental stage, a critical event in establishing the left-right asymmetry of gustatory neurons in C. elegans 107,108. What is emerging is the importance of understanding how cell type and developmental stage-specific changes in genome architecture intersect with the timely expression of key cell fate determinants in order to specify the correct cell fate. Another fascinating direction for future research would be to determine whether Type II neuroblasts and/or INPs also undergo competence transitions. Reorganization of the Type II neuroblast genome, as observed in embryonic neuroblasts 17, could explain how the same set of temporal identity factors in the INPs can give rise to distinct neural identities as the parental Type II neuroblast ages over time 24. Perhaps regulation of genome architecture is a general mechanism that allows a limited group of transcriptional regulators to generate cellular diversity in a spatially and temporally controlled manner.
Changing progenitor competence in mammals
The first examples of changes in competence states of neural progenitors during development came from studies in the mammalian cortex and retina. A series of elegant studies used heterochronic transplantation experiments in the ferret to expose young or old neural progenitors to the opposite host environment and probe for their ability to generate host-appropriate laminar fates 109,110. Early cortical progenitors, which normally produce deep-layer V and VI neurons, were competent to produce the later-born, superficial layer II-IV neurons when transplanted into an older embryo, but not vice versa 109 FIGURE 6A. Remarkably, when early cortical progenitors were transplanted during S-phase, or exposed to brain-derived neurotrophic factor prior to S-phase, they were competent to follow the host program and produce late-born neurons 111,112. In contrast, older progenitors were not competent to generate deep-layer, early-born neurons even if they had undergone one or more rounds of cell division in the younger host environment 109. Interestingly, when layer IV progenitors were heterochronically transplanted into a younger host environment making layer VI neurons, the donor progenitors were no longer competent to produce the early-born layer VI neurons, but were still able to give rise to layer V neurons 113. Since the donor progenitors were isolated after layer V neuron production had already ceased, the results suggest that competence to specify temporal identity persists for a limited time after the generation of that cell type and is not governed by counting cell divisions. This result is strikingly reminiscent of Drosophila neuroblasts, which remain competent to specify early-born identity for several divisions after these cell types cease to be produced and highlight a fundamental property of neural stem cells that competence transitions are not temporally aligned with cell fate transitions.
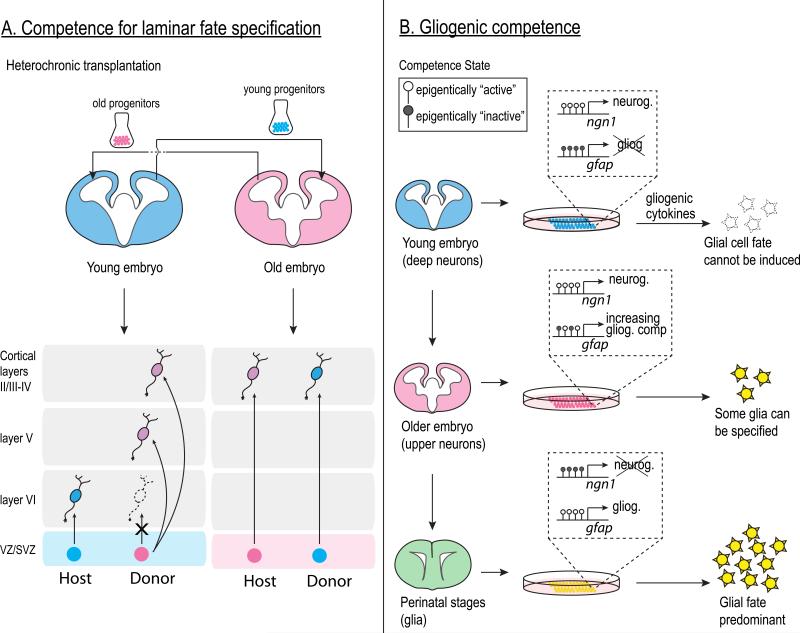
Figure 6. Competence transitions during mammalian neurogenesis.
A. Cortical progenitors lose competence to specify early-born neuronal phenotypes over time. Heterochronic transplantation experiments, in which neural progenitors isolated from one developmental stage (donor) is placed in a similar environment of a different stage (host), show that early progenitors (blue) transplanted into an older host can give rise to later-born phenotypes (red); however older progenitors (red) transplanted into the young embryo do not give rise to early born (layer VI) phenotypes.
B. Changes in chromatin structure at neuronal and gliogenic genes as development progresses contribute to the neurogenic to gliogenic competence transition in the embryo. In early progenitors, regulatory DNA sequences of key gliogenic genes (such as GFAP) are hypermethylated and silenced. In older progenitors, these DNA regions become hypomethylated and are now competent for transcriptional activation. Neural progenitors cultured from older embryos (red and green embryos) are more competent to respond to gliogenic signals to give rise to glial cells (green stars).
The identification of mammalian temporal identity factors has facilitated investigation of mammalian neural progenitor competence. When Ikaros is ectopically expressed in older retinal progenitors, it can induce production of early-born neuronal identities, like horizontal and amacrine cells, and suppress the production of the late-born Muller glia 51. However, Ikaros misexpression could not generate early-born ganglion cells in vivo, suggesting that some but not all early progenitor competence can be restored 51. Similarly, when Lin28 mRNA, a late retinal progenitor microRNA target, is ectopically expressed in early progenitors, there is an increase in the Brn3+ early-born ganglion cell type, but not when expressed in late progenitors, suggesting a limited competence window to specify early-born fate 94. In the cortex, ectopic expression of Fezf2 in late cortical progenitors induces production of neuronal progeny with characteristics of early-born neurons 75. However, the neurons still migrate to superficial layers and were able to make callosal projections, characteristic of late-born neurons, suggesting that older progenitors are not fully competent to specify early-born identity. More recently, competence to respond to Fezf2 was studied in postmitotic neurons, revealing that ectopic expression of Fezf2 in postmitotic, upper cortical neurons can reprogram them to adopt characteristics of deep layer neurons, including the expression of the appropriate molecular markers, axonal projection patterns, and physiological phenotypes 114,115. Interestingly, these postmitotic neurons could switch phenotypes only for a brief time window after their terminal mitosis, suggesting a progressive restriction in competence as the postmitotic neurons aged. Work in Drosophila has previously shown that late-born postmitotic neurons are not competent to adopt early-born neuron characteristics upon misexpression of Hb 7, but it is not clear whether loss of competence occurs immediately upon neuronal birth or whether there exists a short competence period to switch fates. It will be important to determine what aspects of competence become restricted over time in the progenitor versus the progeny.
Culturing neural progenitors in vitro has provided information on the role of extrinsic cues in regulating progenitor competence. Pioneering work showed that rat retinal progenitors dissociated in vitro generate progeny on the same schedule as they do in vivo. Moreover, when co-cultured with progenitors from a different developmental stage they can respond to diffusible extrinsic signals and alter the proportions of progeny subtypes produced, but not the timing 116,117. In fact, when old progenitors are cultured in an excess of young progenitors (or vice versa), changes in environmental signals can bias the relative proportion of cell fates generated, but cannot induce them to specify cell fates outside of their temporal window 118-120, suggesting that retinal progenitors pass through multiple competence stages over the course of their lineage (reviewed in 121). Interestingly, ectopic expression of Ikaros in late-stage retinal progenitors can induce transcription of the early-born ganglion cell marker, Brn3, only when progenitors are cultured in vitro, but not in vivo 51, suggesting a role for extrinsic signals in terminating an early competence window. Similar to the retina, mammalian cortical progenitors in vitro can also sequentially produce neurons with gene expression appropriate for their birth order, and they even lose competence to specify earlier-born neuronal fates 122. Together, observations from retina and cortical studies implicate a coordination of cell-intrinsic timing mechanisms and extrinsic signals that regulate competence.
The switch from neurogenesis to gliogenesis occurs in multiple regions of the developing CNS and reflects an important competence transition. Neurogenic cortical progenitors cultured on embryonic brain slices make neurons but can switch to making glia when cultured on postnatal brain slices123. Consistently, ectopic expression of CNTF in vivo can induce neurogenic progenitors to precociously produce glia 84. These results suggest that at least a subset of neurogenic progenitors are competent to make glia when exposed to the proper extrinsic signals. However, early cortical progenitors have limited competence to generate glia, as suggested by the lack of glia produced during early corticogenesis despite the presence of gliogenic cytokines124,125, and young cortical progenitors generated fewer glial cells than older progenitors when exposed to gliogenic cytokines in culture 126 FIGURE 6B. This difference in gliogenic competence is due at least in part to the highly methylated status of the GFAP promoter during the neurogenic phase, as revealed by the finding that neural progenitors lacking the DNA methyltransferase, Dnmt1 can precociously generate GFAP+ astrocytes in response to LIF 127,128. COUP-TFI/II, which are transiently expressed just before the onset of gliogenesis, are required to release silencing chromatin modifications at promoter regions of glial genes and allow progenitors to become gliogenically competent 89. The Polycomb group complex has also been found to silence the proneural bHLH genes to end neurogenesis 129, highlighting the role of epigenetic mechanisms in the competence switch. Interestingly, a recent study observed that the switch from neurogenesis to gliogenesis involves a change in the progenitor competence that is independent from the progenitor temporal identity transition. Sustained expression of Ikaros, an early-born temporal identity factor during cortical neurogenesis, dramatically affects the fate of the neuronal progeny, but does not affect the timing of gliogenesis 79 FIGURE 4C - neurons adopt the early-born, deep layer phenotypes at the expense of the later-born, upper layer phenotypes, but gliogenesis still begins on time. These results are strikingly similar to those observed in Drosophila neuroblasts, where maintained expression of Hb produces ectopic neurons with early-born identity only during the early competence window. Similarly, Ikaros appears to be able to produce extra early-born neurons in the cortex, but only during the neurogenic competence phase.
Conclusions and Future perspectives
We are only scratching the surface of understanding how neural stem cell temporal identity and competence are regulated during development, and the work highlighted above reveals new research avenues that would provide insight into this important process. It is clear that the specification of cell fate involves a complex interplay between the expression of temporal identity factors, spatial information, and the competence state of the progenitor. As we have discussed, changes in competence are not necessarily restricted to cell fate decisions during neurogenesis, but can be studied in a broader context of developmental transitions. It is still unclear how mechanisms that regulate competence transitions intersect with changes in progenitor temporal identity. Do unique genome architectures allow the same transcription factors to regulate different target genes appropriate for the cell type and stage? How is neurogenic competence maintained indefinitely in adult neural progenitors? Are these adult progenitors competent to make other types of neural cell types?
What can we learn from studying temporal fate specification and the regulation of progenitor competence to develop more effective tools for tissue repair? How could we start with a pluripotent stem cell and induce specification of a particular cell type? Understanding the mechanism(s) that regulate progenitor competence may help increase the efficiency of somatic cell reprogramming, and understanding how multiple competence windows are established may increase the accuracy of directed cellular reprogramming. These are important and exciting avenues for future research.
Glossary terms
- Neural progenitors
Multipotent progenitors that give rise to the diverse cell types of the central nervous system
- Asymmetric cell division
A mitotic division that generates daughter cells that have different cell fates
- Ganglion mother cell
the differentiating daughter cell derived from a neuroblast's asymmetric division. This intermediate progenitor cell will divide once more to generate two neurons or glia.
- Cytokinesis
The final event in the cell-division cycle. Its completion results in the irreversible partition of a mother cell into two daughter cells. It involves cytoplasmic division, driven by an actin-based constriction of the contractile ring.
- Central complex
A region of the fly brain involved in locomotion, vision, learning, and memory.
- Nuclear lamina
a network of intermediate filaments and membrane associated proteins of the nuclear envelope. Genes associated with the nuclear lamina compartment are often in a silenced or repressed state.
- Polycomb Repressive Complexes (PRC)
A multiprotein complex that remodels chromatin to establish epigenetic silencing.
- MicroRNA
a family of short, non-coding RNAs that inhibit translation of mRNAs in a sequence-specific manner.
Literature Cited
- 1.Jessell TM. Neuronal specification in the spinal cord: inductive signals and transcriptional codes. Nature reviews. Genetics. 2000;1:20–29. doi: 10.1038/35049541. doi:10.1038/35049541. [DOI] [PubMed] [Google Scholar]
- 2.Pearson BJ, Doe CQ. Specification of temporal identity in the developing nervous system. Annu Rev Cell Dev Biol. 2004;20:619–647. doi: 10.1146/annurev.cellbio.19.111301.115142. doi:10.1146/annurev.cellbio.19.111301.115142. [DOI] [PubMed] [Google Scholar]
- 3.Broadus J, et al. New neuroblast markers and the origin of the aCC/pCC neurons in the Drosophila central nervous system. Mech Dev. 1995;53:393–402. doi: 10.1016/0925-4773(95)00454-8. doi:0925477395004548 [pii] [DOI] [PubMed] [Google Scholar]
- 4.Doe CQ, Technau GM. Identification and cell lineage of individual neural precursors in the Drosophila CNS. Trends Neurosci. 1993;16:510–514. doi: 10.1016/0166-2236(93)90195-r. [DOI] [PubMed] [Google Scholar]
- 5.Isshiki T, Pearson B, Holbrook S, Doe CQ. Drosophila neuroblasts sequentially express transcription factors which specify the temporal identity of their neuronal progeny. Cell. 2001;106:511–521. doi: 10.1016/s0092-8674(01)00465-2. doi:S0092-8674(01)00465-2 [pii] [DOI] [PubMed] [Google Scholar]
- 6.Bossing T, Udolph G, Doe CQ, Technau GM. The embryonic central nervous system lineages of Drosophila melanogaster. I. Neuroblast lineages derived from the ventral half of the neuroectoderm. Dev Biol. 1996;179:41–64. doi: 10.1006/dbio.1996.0240. doi:S0012-1606(96)90240-7 [pii]10.1006/dbio.1996.0240. [DOI] [PubMed] [Google Scholar]
- 7.Pearson BJ, Doe CQ. Regulation of neuroblast competence in Drosophila. Nature. 2003;425:624–628. doi: 10.1038/nature01910. doi:10.1038/nature01910 nature01910 [pii] [DOI] [PubMed] [Google Scholar]
- 8.Schmid A, Chiba A, Doe CQ. Clonal analysis of Drosophila embryonic neuroblasts: neural cell types, axon projections and muscle targets. Development. 1999;126:4653–4689. doi: 10.1242/dev.126.21.4653. [DOI] [PubMed] [Google Scholar]
- 9.Schmidt H, et al. The embryonic central nervous system lineages of Drosophila melanogaster. II. Neuroblast lineages derived from the dorsal part of the neuroectoderm. Dev Biol. 1997;189:186–204. doi: 10.1006/dbio.1997.8660. doi:S0012160697986607 [pii] [DOI] [PubMed] [Google Scholar]
- 10.Tran KD, Doe CQ. Pdm and Castor close successive temporal identity windows in the NB3-1 lineage. Development. 2008;135:3491–3499. doi: 10.1242/dev.024349. doi:dev.024349 [pii] 10.1242/dev.024349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kambadur R, et al. Regulation of POU genes by castor and hunchback establishes layered compartments in the Drosophila CNS. Genes Dev. 1998;12:246–260. doi: 10.1101/gad.12.2.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cleary MD, Doe CQ. Regulation of neuroblast competence: multiple temporal identity factors specify distinct neuronal fates within a single early competence window. Genes Dev. 2006;20:429–434. doi: 10.1101/gad.1382206. doi:20/4/429 [pii] 10.1101/gad.1382206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Touma JJ, Weckerle FF, Cleary MD. Drosophila Polycomb complexes restrict neuroblast competence to generate motoneurons. Development. 2012;139:657–666. doi: 10.1242/dev.071589. doi:10.1242/dev.071589. [DOI] [PubMed] [Google Scholar]
- 14.Kanai MI, Okabe M, Hiromi Y. seven-up Controls switching of transcription factors that specify temporal identities of Drosophila neuroblasts. Dev Cell. 2005;8:203–213. doi: 10.1016/j.devcel.2004.12.014. doi:10.1016/j.devcel.2004.12.014. [DOI] [PubMed] [Google Scholar]
- 15.Novotny T, Eiselt R, Urban J. Hunchback is required for the specification of the early sublineage of neuroblast 7-3 in the Drosophila central nervous system. Development. 2002;129:1027–1036. doi: 10.1242/dev.129.4.1027. [DOI] [PubMed] [Google Scholar]
- 16.Hirono K, Margolis JS, Posakony JW, Doe CQ. Identification of hunchback cis-regulatory DNA conferring temporal expression in neuroblasts and neurons. Gene expression patterns : GEP. 2012;12:11–17. doi: 10.1016/j.gep.2011.10.001. doi:10.1016/j.gep.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kohwi M, Lupton JR, Lai SL, Miller MR, Doe CQ. Developmentally regulated subnuclear genome reorganization restricts neural progenitor competence in Drosophila. Cell. 2013;152:97–108. doi: 10.1016/j.cell.2012.11.049. doi:10.1016/j.cell.2012.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grosskortenhaus R, Pearson BJ, Marusich A, Doe CQ. Regulation of temporal identity transitions in Drosophila neuroblasts. Dev Cell. 2005;8:193–202. doi: 10.1016/j.devcel.2004.11.019. doi:S1534-5807(04)00456-3 [pii] 10.1016/j.devcel.2004.11.019. [DOI] [PubMed] [Google Scholar]
- 19.Grosskortenhaus R, Robinson KJ, Doe CQ. Pdm and Castor specify late-born motor neuron identity in the NB7-1 lineage. Genes Dev. 2006;20:2618–2627. doi: 10.1101/gad.1445306. doi:20/18/2618 [pii] 10.1101/gad.1445306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baumgardt M, Karlsson D, Terriente J, Diaz-Benjumea FJ, Thor S. Neuronal subtype specification within a lineage by opposing temporal feed-forward loops. Cell. 2009;139:969–982. doi: 10.1016/j.cell.2009.10.032. doi:S0092-8674(09)01358-0 [pii] 10.1016/j.cell.2009.10.032. [DOI] [PubMed] [Google Scholar]
- 21.Brody T, Odenwald WF. Programmed transformations in neuroblast gene expression during Drosophila CNS lineage development. Dev Biol. 2000;226:34–44. doi: 10.1006/dbio.2000.9829. doi:10.1006/dbio.2000.9829 S0012-1606(00)99829-4 [pii] [DOI] [PubMed] [Google Scholar]
- 22.Cenci C, Gould AP. Drosophila Grainyhead specifies late programmes of neural proliferation by regulating the mitotic activity and Hox-dependent apoptosis of neuroblasts. Development. 2005;132:3835–3845. doi: 10.1242/dev.01932. doi:dev.01932 [pii] 10.1242/dev.01932. [DOI] [PubMed] [Google Scholar]
- 23.Maurange C, Cheng L, Gould AP. Temporal transcription factors and their targets schedule the end of neural proliferation in Drosophila. Cell. 2008;133:891–902. doi: 10.1016/j.cell.2008.03.034. doi:S0092-8674(08)00499-6 [pii] 10.1016/j.cell.2008.03.034. [DOI] [PubMed] [Google Scholar]
- 24.Bayraktar OA, Doe CQ. Combinatorial temporal patterning in progenitors expands neural diversity. Nature. 2013;498:449–455. doi: 10.1038/nature12266. doi:10.1038/nature12266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kohwi M, Hiebert LS, Doe CQ. The pipsqueak-domain proteins Distal antenna and Distal antenna-related restrict Hunchback neuroblast expression and early-born neuronal identity. Development. 2011;138:1727–1735. doi: 10.1242/dev.061499. doi:dev.061499 [pii] 10.1242/dev.061499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mettler U, Vogler G, Urban J. Timing of identity: spatiotemporal regulation of hunchback in neuroblast lineages of Drosophila by Seven-up and Prospero. Development. 2006;133:429–437. doi: 10.1242/dev.02229. doi:dev.02229 [pii] 10.1242/dev.02229. [DOI] [PubMed] [Google Scholar]
- 27.Benito-Sipos J, et al. Seven up acts as a temporal factor during two different stages of neuroblast 5-6 development. Development. 2011;138:5311–5320. doi: 10.1242/dev.070946. doi:10.1242/dev.070946. [DOI] [PubMed] [Google Scholar]
- 28.Kao CF, Yu HH, He Y, Kao JC, Lee T. Hierarchical deployment of factors regulating temporal fate in a diverse neuronal lineage of the Drosophila central brain. Neuron. 2012;73:677–684. doi: 10.1016/j.neuron.2011.12.018. doi:10.1016/j.neuron.2011.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhu S, et al. Gradients of the Drosophila Chinmo BTB-zinc finger protein govern neuronal temporal identity. Cell. 2006;127:409–422. doi: 10.1016/j.cell.2006.08.045. doi:S0092-8674(06)01230-X [pii] 10.1016/j.cell.2006.08.045. [DOI] [PubMed] [Google Scholar]
- 30.Wu YC, Chen CH, Mercer A, Sokol NS. Let-7-complex microRNAs regulate the temporal identity of Drosophila mushroom body neurons via chinmo. Dev Cell. 2012;23:202–209. doi: 10.1016/j.devcel.2012.05.013. doi:10.1016/j.devcel.2012.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Egger B, Boone JQ, Stevens NR, Brand AH, Doe CQ. Regulation of spindle orientation and neural stem cell fate in the Drosophila optic lobe. Neural Dev. 2007;2:1. doi: 10.1186/1749-8104-2-1. doi:1749-8104-2-1 [pii] 10.1186/1749-8104-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Egger B, Gold KS, Brand AH. Notch regulates the switch from symmetric to asymmetric neural stem cell division in the Drosophila optic lobe. Development. 2010;137:2981–2987. doi: 10.1242/dev.051250. doi:10.1242/dev.051250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yasugi T, Umetsu D, Murakami S, Sato M, Tabata T. Drosophila optic lobe neuroblasts triggered by a wave of proneural gene expression that is negatively regulated by JAK/STAT. Development. 2008;135:1471–1480. doi: 10.1242/dev.019117. doi:10.1242/dev.019117. [DOI] [PubMed] [Google Scholar]
- 34.Li X, et al. Temporal patterning of Drosophila medulla neuroblasts controls neural fates. Nature. 2013 doi: 10.1038/nature12319. doi:10.1038/nature12319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suzuki T, Kaido M, Takayama R, Sato M. A temporal mechanism that produces neuronal diversity in the Drosophila visual center. Dev Biol. 2013;380:12–24. doi: 10.1016/j.ydbio.2013.05.002. doi:10.1016/j.ydbio.2013.05.002. [DOI] [PubMed] [Google Scholar]
- 36.Bello BC, Izergina N, Caussinus E, Reichert H. Amplification of neural stem cell proliferation by intermediate progenitor cells in Drosophila brain development. Neural Dev. 2008;3:5. doi: 10.1186/1749-8104-3-5. doi:1749-8104-3-5 [pii] 10.1186/1749-8104-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boone JQ, Doe CQ. Identification of Drosophila type II neuroblast lineages containing transit amplifying ganglion mother cells. Dev Neurobiol. 2008;68:1185–1195. doi: 10.1002/dneu.20648. doi:10.1002/dneu.20648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bowman SK, et al. The tumor suppressors Brat and Numb regulate transit-amplifying neuroblast lineages in Drosophila. Dev Cell. 2008;14:535–546. doi: 10.1016/j.devcel.2008.03.004. doi:S1534-5807(08)00112-3 [pii] 10.1016/j.devcel.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bayraktar OA, Boone JQ, Drummond ML, Doe CQ. Drosophila type II neuroblast lineages keep Prospero levels low to generate large clones that contribute to the adult brain central complex. Neural Dev. 2010;5:26. doi: 10.1186/1749-8104-5-26. doi:1749-8104-5-26 [pii] 10.1186/1749-8104-5-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bayraktar OA, Doe CQ. Combinatorial temporal patterning in progenitors expands neural diversity. Nature. 2013 doi: 10.1038/nature12266. doi:10.1038/nature12266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Izergina N, Balmer J, Bello B, Reichert H. Postembryonic development of transit amplifying neuroblast lineages in the Drosophila brain. Neural Dev. 2009;4:44. doi: 10.1186/1749-8104-4-44. doi:1749-8104-4-44 [pii] 10.1186/1749-8104-4-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Doetsch F, Caille I, Lim DA, Garcia-Verdugo JM, Alvarez-Buylla A. Subventricular zone astrocytes are neural stem cells in the adult mammalian brain. Cell. 1999;97:703–716. doi: 10.1016/s0092-8674(00)80783-7. [DOI] [PubMed] [Google Scholar]
- 43.Hansen DV, Lui JH, Parker PR, Kriegstein AR. Neurogenic radial glia in the outer subventricular zone of human neocortex. Nature. 2010;464:554–561. doi: 10.1038/nature08845. doi:nature08845 [pii] 10.1038/nature08845. [DOI] [PubMed] [Google Scholar]
- 44.LaMonica BE, Lui JH, Wang X, Kriegstein AR. OSVZ progenitors in the human cortex: an updated perspective on neurodevelopmental disease. Current opinion in neurobiology. 2012;22:747–753. doi: 10.1016/j.conb.2012.03.006. doi:10.1016/j.conb.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fekete DM, Perez-Miguelsanz J, Ryder EF, Cepko CL. Clonal analysis in the chicken retina reveals tangential dispersion of clonally related cells. Dev Biol. 1994;166:666–682. doi: 10.1006/dbio.1994.1346. doi:10.1006/dbio.1994.1346. [DOI] [PubMed] [Google Scholar]
- 46.Wetts R, Fraser SE. Multipotent precursors can give rise to all major cell types of the frog retina. Science. 1988;239:1142–1145. doi: 10.1126/science.2449732. [DOI] [PubMed] [Google Scholar]
- 47.Holt CE, Bertsch TW, Ellis HM, Harris WA. Cellular determination in the Xenopus retina is independent of lineage and birth date. Neuron. 1988;1:15–26. doi: 10.1016/0896-6273(88)90205-x. [DOI] [PubMed] [Google Scholar]
- 48.Turner DL, Cepko CL. A common progenitor for neurons and glia persists in rat retina late in development. Nature. 1987;328:131–136. doi: 10.1038/328131a0. doi:10.1038/328131a0. [DOI] [PubMed] [Google Scholar]
- 49.Turner DL, Snyder EY, Cepko CL. Lineage-independent determination of cell type in the embryonic mouse retina. Neuron. 1990;4:833–845. doi: 10.1016/0896-6273(90)90136-4. [DOI] [PubMed] [Google Scholar]
- 50.Trimarchi JM, Stadler MB, Cepko CL. Individual retinal progenitor cells display extensive heterogeneity of gene expression. PloS one. 2008;3:e1588. doi: 10.1371/journal.pone.0001588. doi:10.1371/journal.pone.0001588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Elliott J, Jolicoeur C, Ramamurthy V, Cayouette M. Ikaros confers early temporal competence to mouse retinal progenitor cells. Neuron. 2008;60:26–39. doi: 10.1016/j.neuron.2008.08.008. doi:S0896-6273(08)00676-4 [pii] 10.1016/j.neuron.2008.08.008. [DOI] [PubMed] [Google Scholar]
- 52.Gomes FL, et al. Reconstruction of rat retinal progenitor cell lineages in vitro reveals a surprising degree of stochasticity in cell fate decisions. Development. 2011;138:227–235. doi: 10.1242/dev.059683. doi:10.1242/dev.059683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Johnston RJ, Jr., Desplan C. Stochastic mechanisms of cell fate specification that yield random or robust outcomes. Annual review of cell and developmental biology. 2010;26:689–719. doi: 10.1146/annurev-cellbio-100109-104113. doi:10.1146/annurev cellbio-100109-104113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.De la Huerta I, Kim IJ, Voinescu PE, Sanes JR. Direction-selective retinal ganglion cells arise from molecularly specified multipotential progenitors. Proc Natl Acad Sci U S A. 2012;109:17663–17668. doi: 10.1073/pnas.1215806109. doi:10.1073/pnas.1215806109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hafler BP, et al. Transcription factor Olig2 defines subpopulations of retinal progenitor cells biased toward specific cell fates. Proc Natl Acad Sci U S A. 2012;109:7882–7887. doi: 10.1073/pnas.1203138109. doi:10.1073/pnas.1203138109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Leone DP, Srinivasan K, Chen B, Alcamo E, McConnell SK. The determination of projection neuron identity in the developing cerebral cortex. Current opinion in neurobiology. 2008;18:28–35. doi: 10.1016/j.conb.2008.05.006. doi:10.1016/j.conb.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Molyneaux BJ, Arlotta P, Menezes JR, Macklis JD. Neuronal subtype specification in the cerebral cortex. Nature reviews. Neuroscience. 2007;8:427–437. doi: 10.1038/nrn2151. doi:10.1038/nrn2151. [DOI] [PubMed] [Google Scholar]
- 58.Noctor SC, Martinez-Cerdeno V, Ivic L, Kriegstein AR. Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat Neurosci. 2004;7:136–144. doi: 10.1038/nn1172. [DOI] [PubMed] [Google Scholar]
- 59.Noctor SC, Martinez-Cerdeno V, Kriegstein AR. Distinct behaviors of neural stem and progenitor cells underlie cortical neurogenesis. The Journal of comparative neurology. 2008;508:28–44. doi: 10.1002/cne.21669. doi:10.1002/cne.21669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gotz M, Stoykova A, Gruss P. Pax6 controls radial glia differentiation in the cerebral cortex. Neuron. 1998;21:1031–1044. doi: 10.1016/s0896-6273(00)80621-2. [DOI] [PubMed] [Google Scholar]
- 61.Nieto M, et al. Expression of Cux-1 and Cux-2 in the subventricular zone and upper layers II-IV of the cerebral cortex. The Journal of comparative neurology. 2004;479:168–180. doi: 10.1002/cne.20322. doi:10.1002/cne.20322. [DOI] [PubMed] [Google Scholar]
- 62.Tarabykin V, Stoykova A, Usman N, Gruss P. Cortical upper layer neurons derive from the subventricular zone as indicated by Svet1 gene expression. Development. 2001;128:1983–1993. doi: 10.1242/dev.128.11.1983. [DOI] [PubMed] [Google Scholar]
- 63.Zimmer C, Tiveron MC, Bodmer R, Cremer H. Dynamics of Cux2 expression suggests that an early pool of SVZ precursors is fated to become upper cortical layer neurons. Cerebral cortex. 2004;14:1408–1420. doi: 10.1093/cercor/bhh102. doi:10.1093/cercor/bhh102. [DOI] [PubMed] [Google Scholar]
- 64.Franco SJ, et al. Fate-restricted neural progenitors in the mammalian cerebral cortex. Science. 2012;337:746–749. doi: 10.1126/science.1223616. doi:10.1126/science.1223616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yu YC, Bultje RS, Wang X, Shi SH. Specific synapses develop preferentially among sister excitatory neurons in the neocortex. Nature. 2009;458:501–504. doi: 10.1038/nature07722. doi:10.1038/nature07722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cubelos B, et al. Cux-2 controls the proliferation of neuronal intermediate precursors of the cortical subventricular zone. Cerebral cortex. 2008;18:1758–1770. doi: 10.1093/cercor/bhm199. doi:10.1093/cercor/bhm199. [DOI] [PubMed] [Google Scholar]
- 67.Harrison SJ, Nishinakamura R, Jones KR, Monaghan AP. Sall1 regulates cortical neurogenesis and laminar fate specification in mice: implications for neural abnormalities in Townes- Brocks syndrome. Disease models & mechanisms. 2012;5:351–365. doi: 10.1242/dmm.002873. doi:10.1242/dmm.002873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rodriguez M, Choi J, Park S, Sockanathan S. Gde2 regulates cortical neuronal identity by controlling the timing of cortical progenitor differentiation. Development. 2012;139:3870–3879. doi: 10.1242/dev.081083. doi:10.1242/dev.081083. [DOI] [PubMed] [Google Scholar]
- 69.Decembrini S, et al. MicroRNAs couple cell fate and developmental timing in retina. Proc Natl Acad Sci U S A. 2009;106:21179–21184. doi: 10.1073/pnas.0909167106. doi:10.1073/pnas.0909167106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Saade M, et al. Sonic hedgehog signaling switches the mode of division in the developing nervous system. Cell Rep. 2013;4:492–503. doi: 10.1016/j.celrep.2013.06.038. doi:10.1016/j.celrep.2013.06.038. [DOI] [PubMed] [Google Scholar]
- 71.Alcamo EA, et al. Satb2 regulates callosal projection neuron identity in the developing cerebral cortex. Neuron. 2008;57:364–377. doi: 10.1016/j.neuron.2007.12.012. doi:10.1016/j.neuron.2007.12.012. [DOI] [PubMed] [Google Scholar]
- 72.Arlotta P, Molyneaux BJ, Jabaudon D, Yoshida Y, Macklis JD. Ctip2 controls the differentiation of medium spiny neurons and the establishment of the cellular architecture of the striatum. J Neurosci. 2008;28:622–632. doi: 10.1523/JNEUROSCI.2986-07.2008. doi:10.1523/JNEUROSCI.2986-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen B, Schaevitz LR, McConnell SK. Fezl regulates the differentiation and axon targeting of layer 5 subcortical projection neurons in cerebral cortex. Proc Natl Acad Sci U S A. 2005;102:17184–17189. doi: 10.1073/pnas.0508732102. doi:10.1073/pnas.0508732102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chen B, et al. The Fezf2-Ctip2 genetic pathway regulates the fate choice of subcortical projection neurons in the developing cerebral cortex. Proc Natl Acad Sci U S A. 2008;105:11382–11387. doi: 10.1073/pnas.0804918105. doi:10.1073/pnas.0804918105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chen JG, Rasin MR, Kwan KY, Sestan N. Zfp312 is required for subcortical axonal projections and dendritic morphology of deep-layer pyramidal neurons of the cerebral cortex. Proc Natl Acad Sci U S A. 2005;102:17792–17797. doi: 10.1073/pnas.0509032102. doi:10.1073/pnas.0509032102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Britanova O, et al. Satb2 is a postmitotic determinant for upper-layer neuron specification in the neocortex. Neuron. 2008;57:378–392. doi: 10.1016/j.neuron.2007.12.028. doi:10.1016/j.neuron.2007.12.028. [DOI] [PubMed] [Google Scholar]
- 77.Sugitani Y, et al. Brn-1 and Brn-2 share crucial roles in the production and positioning of mouse neocortical neurons. Genes & development. 2002;16:1760–1765. doi: 10.1101/gad.978002. doi:10.1101/gad.978002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Srinivasan K, et al. A network of genetic repression and derepression specifies projection fates in the developing neocortex. Proc Natl Acad Sci U S A. 2012;109:19071–19078. doi: 10.1073/pnas.1216793109. doi:10.1073/pnas.1216793109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Alsio JM, Tarchini B, Cayouette M, Livesey FJ. Ikaros promotes early-born neuronal fates in the cerebral cortex. Proc Natl Acad Sci U S A. 2013;110:E716–725. doi: 10.1073/pnas.1215707110. doi:10.1073/pnas.1215707110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Guillemot F. Cell fate specification in the mammalian telencephalon. Prog Neurobiol. 2007;83:37–52. doi: 10.1016/j.pneurobio.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 81.Rowitch DH, Kriegstein AR. Developmental genetics of vertebrate glial-cell specification. Nature. 2010;468:214–222. doi: 10.1038/nature09611. doi:10.1038/nature09611. [DOI] [PubMed] [Google Scholar]
- 82.Ross SE, Greenberg ME, Stiles CD. Basic helix-loop-helix factors in cortical development. Neuron. 2003;39:13–25. doi: 10.1016/s0896-6273(03)00365-9. [DOI] [PubMed] [Google Scholar]
- 83.Stolt CC, et al. The Sox9 transcription factor determines glial fate choice in the developing spinal cord. Genes & development. 2003;17:1677–1689. doi: 10.1101/gad.259003. doi:10.1101/gad.259003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Barnabe-Heider F, et al. Evidence that embryonic neurons regulate the onset of cortical gliogenesis via cardiotrophin-1. Neuron. 2005;48:253–265. doi: 10.1016/j.neuron.2005.08.037. doi:10.1016/j.neuron.2005.08.037. [DOI] [PubMed] [Google Scholar]
- 85.Bonni A, et al. Regulation of gliogenesis in the central nervous system by the JAK-STAT signaling pathway. Science. 1997;278:477–483. doi: 10.1126/science.278.5337.477. [DOI] [PubMed] [Google Scholar]
- 86.Koblar SA, et al. Neural precursor differentiation into astrocytes requires signaling through the leukemia inhibitory factor receptor. Proc Natl Acad Sci U S A. 1998;95:3178–3181. doi: 10.1073/pnas.95.6.3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nakashima K, et al. Developmental requirement of gp130 signaling in neuronal survival and astrocyte differentiation. J Neurosci. 1999;19:5429–5434. doi: 10.1523/JNEUROSCI.19-13-05429.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Miller FD, Gauthier AS. Timing is everything: making neurons versus glia in the developing cortex. Neuron. 2007;54:357–369. doi: 10.1016/j.neuron.2007.04.019. doi:10.1016/j.neuron.2007.04.019. [DOI] [PubMed] [Google Scholar]
- 89.Naka H, Nakamura S, Shimazaki T, Okano H. Requirement for COUP-TFI and II in the temporal specification of neural stem cells in CNS development. Nat Neurosci. 2008;11:1014–1023. doi: 10.1038/nn.2168. doi:10.1038/nn.2168. [DOI] [PubMed] [Google Scholar]
- 90.Faedo A, et al. COUP-TFI coordinates cortical patterning, neurogenesis, and laminar fate and modulates MAPK/ERK, AKT, and beta-catenin signaling. Cerebral cortex. 2008;18:2117–2131. doi: 10.1093/cercor/bhm238. doi:10.1093/cercor/bhm238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.De Pietri Tonelli D, et al. miRNAs are essential for survival and differentiation of newborn neurons but not for expansion of neural progenitors during early neurogenesis in the mouse embryonic neocortex. Development. 2008;135:3911–3921. doi: 10.1242/dev.025080. doi:10.1242/dev.025080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Georgi SA, Reh TA. Dicer is required for the transition from early to late progenitor state in the developing mouse retina. J Neurosci. 2010;30:4048–4061. doi: 10.1523/JNEUROSCI.4982-09.2010. doi:10.1523/JNEUROSCI.4982-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kawase-Koga Y, Otaegi G, Sun T. Different timings of Dicer deletion affect neurogenesis and gliogenesis in the developing mouse central nervous system. Dev Dyn. 2009;238:2800–2812. doi: 10.1002/dvdy.22109. doi:10.1002/dvdy.22109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.La Torre A, Georgi S, Reh TA. Conserved microRNA pathway regulates developmental timing of retinal neurogenesis. Proc Natl Acad Sci U S A. 2013 doi: 10.1073/pnas.1301837110. doi:10.1073/pnas.1301837110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cremisi F. MicroRNAs and cell fate in cortical and retinal development. Front Cell Neurosci. 2013;7:141. doi: 10.3389/fncel.2013.00141. doi:10.3389/fncel.2013.00141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zheng K, Li H, Zhu Y, Zhu Q, Qiu M. MicroRNAs are essential for the developmental switch from neurogenesis to gliogenesis in the developing spinal cord. J Neurosci. 2010;30:8245–8250. doi: 10.1523/JNEUROSCI.1169-10.2010. doi:10.1523/JNEUROSCI.1169-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Moss EG. Heterochronic genes and the nature of developmental time. Current biology : CB. 2007;17:R425–434. doi: 10.1016/j.cub.2007.03.043. doi:10.1016/j.cub.2007.03.043. [DOI] [PubMed] [Google Scholar]
- 98.Abbott AL, et al. The let-7 MicroRNA family members mir-48, mir-84, and mir-241 function together to regulate developmental timing in Caenorhabditis elegans. Dev Cell. 2005;9:403–414. doi: 10.1016/j.devcel.2005.07.009. doi:10.1016/j.devcel.2005.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Davis TH, et al. Conditional loss of Dicer disrupts cellular and tissue morphogenesis in the cortex and hippocampus. J Neurosci. 2008;28:4322–4330. doi: 10.1523/JNEUROSCI.4815-07.2008. doi:10.1523/JNEUROSCI.4815-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Meister P, Mango SE, Gasser SM. Locking the genome: nuclear organization and cell fate. Current opinion in genetics & development. 2011;21:167–174. doi: 10.1016/j.gde.2011.01.023. doi:10.1016/j.gde.2011.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Peric-Hupkes D, van Steensel B. Role of the nuclear lamina in genome organization and gene expression. Cold Spring Harb Symp Quant Biol. 2010;75:517–524. doi: 10.1101/sqb.2010.75.014. doi:sqb.2010.75.014 [pii]10.1101/sqb.2010.75.014. [DOI] [PubMed] [Google Scholar]
- 102.Shevelyov YY, Nurminsky DI. The nuclear lamina as a gene-silencing hub. Current issues in molecular biology. 2012;14:27–38. [PubMed] [Google Scholar]
- 103.Zhao R, Bodnar MS, Spector DL. Nuclear neighborhoods and gene expression. Current opinion in genetics & development. 2009;19:172–179. doi: 10.1016/j.gde.2009.02.007. doi:10.1016/j.gde.2009.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Siegmund T, Lehmann M. The Drosophila Pipsqueak protein defines a new family of helix-turn-helix DNA-binding proteins. Dev Genes Evol. 2002;212:152–157. doi: 10.1007/s00427-002-0219-2. doi:10.1007/s00427-002-0219-2. [DOI] [PubMed] [Google Scholar]
- 105.Broadus J, Doe CQ. Evolution of neuroblast identity: seven-up and prospero expression reveal homologous and divergent neuroblast fates in Drosophila and Schistocerca. Development. 1995;121:3989–3996. doi: 10.1242/dev.121.12.3989. [DOI] [PubMed] [Google Scholar]
- 106.Schwartz YB, Pirrotta V. Polycomb complexes and epigenetic states. Curr Opin Cell Biol. 2008;20:266–273. doi: 10.1016/j.ceb.2008.03.002. doi:10.1016/j.ceb.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 107.Cochella L, Hobert O. Embryonic priming of a miRNA locus predetermines postmitotic neuronal left/right asymmetry in C. elegans. Cell. 2012;151:1229–1242. doi: 10.1016/j.cell.2012.10.049. doi:10.1016/j.cell.2012.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Tursun B, Patel T, Kratsios P, Hobert O. Direct conversion of C. elegans germ cells into specific neuron types. Science. 2011;331:304–308. doi: 10.1126/science.1199082. doi:10.1126/science.1199082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Frantz GD, McConnell SK. Restriction of late cerebral cortical progenitors to an upper-layer fate. Neuron. 1996;17:55–61. doi: 10.1016/s0896-6273(00)80280-9. [DOI] [PubMed] [Google Scholar]
- 110.McConnell SK. Fates of visual cortical neurons in the ferret after isochronic and heterochronic transplantation. J Neurosci. 1988;8:945–974. doi: 10.1523/JNEUROSCI.08-03-00945.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.McConnell SK, Kaznowski CE. Cell cycle dependence of laminar determination in developing neocortex. Science. 1991;254:282–285. doi: 10.1126/science.254.5029.282. [DOI] [PubMed] [Google Scholar]
- 112.Fukumitsu H, et al. Brain-derived neurotrophic factor participates in determination of neuronal laminar fate in the developing mouse cerebral cortex. J Neurosci. 2006;26:13218–13230. doi: 10.1523/JNEUROSCI.4251-06.2006. doi:10.1523/JNEUROSCI.4251-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Desai AR, McConnell SK. Progressive restriction in fate potential by neural progenitors during cerebral cortical development. Development. 2000;127:2863–2872. doi: 10.1242/dev.127.13.2863. [DOI] [PubMed] [Google Scholar]
- 114.De la Rossa A, et al. In vivo reprogramming of circuit connectivity in postmitotic neocortical neurons. Nat Neurosci. 2013;16:193–200. doi: 10.1038/nn.3299. doi:10.1038/nn.3299. [DOI] [PubMed] [Google Scholar]
- 115.Rouaux C, Arlotta P. Direct lineage reprogramming of post-mitotic callosal neurons into corticofugal neurons in vivo. Nature cell biology. 2013;15:214–221. doi: 10.1038/ncb2660. doi:10.1038/ncb2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Watanabe T, Raff MC. Rod photoreceptor development in vitro: intrinsic properties of proliferating neuroepithelial cells change as development proceeds in the rat retina. Neuron. 1990;4:461–467. doi: 10.1016/0896-6273(90)90058-n. [DOI] [PubMed] [Google Scholar]
- 117.Watanabe T, Raff MC. Diffusible rod-promoting signals in the developing rat retina. Development. 1992;114:899–906. doi: 10.1242/dev.114.4.899. [DOI] [PubMed] [Google Scholar]
- 118.Altshuler D, Cepko C. A temporally regulated, diffusible activity is required for rod photoreceptor development in vitro. Development. 1992;114:947–957. doi: 10.1242/dev.114.4.947. [DOI] [PubMed] [Google Scholar]
- 119.Belliveau MJ, Cepko CL. Extrinsic and intrinsic factors control the genesis of amacrine and cone cells in the rat retina. Development. 1999;126:555–566. doi: 10.1242/dev.126.3.555. [DOI] [PubMed] [Google Scholar]
- 120.Belliveau MJ, Young TL, Cepko CL. Late retinal progenitor cells show intrinsic limitations in the production of cell types and the kinetics of opsin synthesis. J Neurosci. 2000;20:2247–2254. doi: 10.1523/JNEUROSCI.20-06-02247.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Livesey FJ, Cepko CL. Vertebrate neural cell-fate determination: lessons from the retina. Nat Rev Neurosci. 2001;2:109–118. doi: 10.1038/35053522. doi:10.1038/35053522. [DOI] [PubMed] [Google Scholar]
- 122.Shen Q, et al. The timing of cortical neurogenesis is encoded within lineages of individual progenitor cells. Nat Neurosci. 2006;9:743–751. doi: 10.1038/nn1694. doi:10.1038/nn1694. [DOI] [PubMed] [Google Scholar]
- 123.Morrow EM, Chen CM, Cepko CL. Temporal order of bipolar cell genesis in the neural retina. Neural Dev. 2008;3:2. doi: 10.1186/1749-8104-3-2. doi:1749-8104-3-2 [pii] 10.1186/1749-8104-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Derouet D, et al. Neuropoietin, a new IL-6-related cytokine signaling through the ciliary neurotrophic factor receptor. Proc Natl Acad Sci U S A. 2004;101:4827–4832. doi: 10.1073/pnas.0306178101. doi:10.1073/pnas.0306178101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Uemura A, et al. Cardiotrophin-like cytokine induces astrocyte differentiation of fetal neuroepithelial cells via activation of STAT3. Cytokine. 2002;18:1–7. doi: 10.1006/cyto.2002.1006. [DOI] [PubMed] [Google Scholar]
- 126.He F, et al. A positive autoregulatory loop of Jak-STAT signaling controls the onset of astrogliogenesis. Nat Neurosci. 2005;8:616–625. doi: 10.1038/nn1440. doi:10.1038/nn1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Takizawa T, et al. DNA methylation is a critical cell-intrinsic determinant of astrocyte differentiation in the fetal brain. Dev Cell. 2001;1:749–758. doi: 10.1016/s1534-5807(01)00101-0. doi:S1534-5807(01)00101-0 [pii] [DOI] [PubMed] [Google Scholar]
- 128.Fan G, et al. DNA methylation controls the timing of astrogliogenesis through regulation of JAK-STAT signaling. Development. 2005;132:3345–3356. doi: 10.1242/dev.01912. doi:10.1242/dev.01912. [DOI] [PubMed] [Google Scholar]
- 129.Hirabayashi Y, et al. Polycomb limits the neurogenic competence of neural precursor cells to promote astrogenic fate transition. Neuron. 2009;63:600–613. doi: 10.1016/j.neuron.2009.08.021. doi:10.1016/j.neuron.2009.08.021. [DOI] [PubMed] [Google Scholar]
- 130.Wurst W, Auerbach AB, Joyner AL. Multiple developmental defects in Engrailed-1 mutant mice: an early mid-hindbrain deletion and patterning defects in forelimbs and sternum. Development. 1994;120:2065–2075. doi: 10.1242/dev.120.7.2065. [DOI] [PubMed] [Google Scholar]