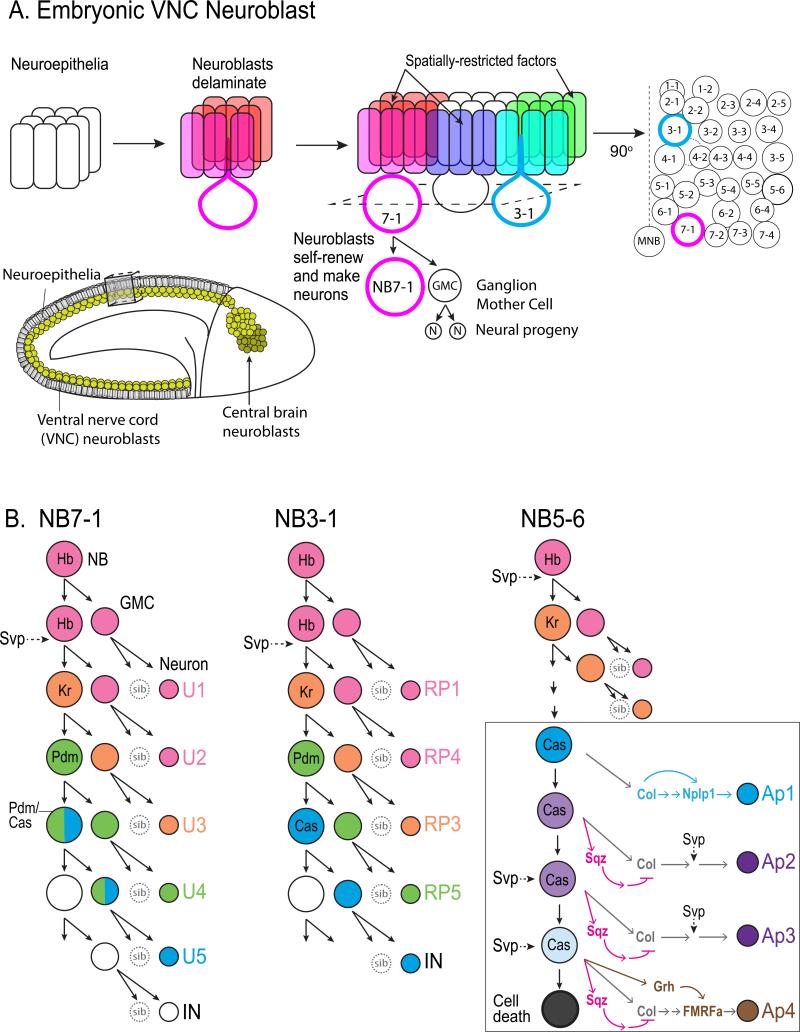
Figure 1. Neurogenesis in Drosophila embryo ventral nerve cord neuroblast lineages.
A. (a1) Neuroblasts are specified and subsequently extruded (delaminated) from the neuroepithelia at the onset of neurogenesis. (a2) Each individual neuroblast can be uniquely identified based on its stereotyped position within the nerve cord and its expression of spatially restricted factors. (a3) Neuroblasts are named according to their row and column within the neuroblast “grid.” Thus, NB7-1 (red) is present in the seventh row and occupies the first column position, whereas NB3-1 (blue) is positioned in the third row. (a4) Each neuroblast undergoes a series of asymmetric divisions that give rise to a self-renewed neuroblast and a differentiating ganglion mother cell (GMC). The GMC divides again to generate two neural progeny. The inset shows a Drosophila embryo at stage 9, at the onset of neuroblast delamination. Neuroblasts are shown in yellow along the ventral nerve cord just beneath epithelial layer. b-d. The lineages of NB7-1, NB3-1, and NB5-6 neuroblasts are shown. Each sequentially expresses the temporal identity factors Hunchback (Hb), Kruppel (Kr), Pou domain protein (Pdm), and Castor (Cas) and give rise to a unique lineage of neural progeny in a stereotyped birth order. The COUP-family nuclear receptor, Seven-up (Svp) is transiently expressed in neuroblasts to regulate timing of temporal fate determinant expression. D, inset. At the end of the NB5-6 lineage, Castor initiates a subtemporal transcriptional cascade by activating Squeeze (Sqz, red rectangle) and Grainyhead (Grh, green rectangle). The delay in the accumulation of Sqz and Grh as NB5-6 divides results in distinct neuronal subtypes generated during the Castor window. At the end of the lineage, NB5-6 undergoes cell death.