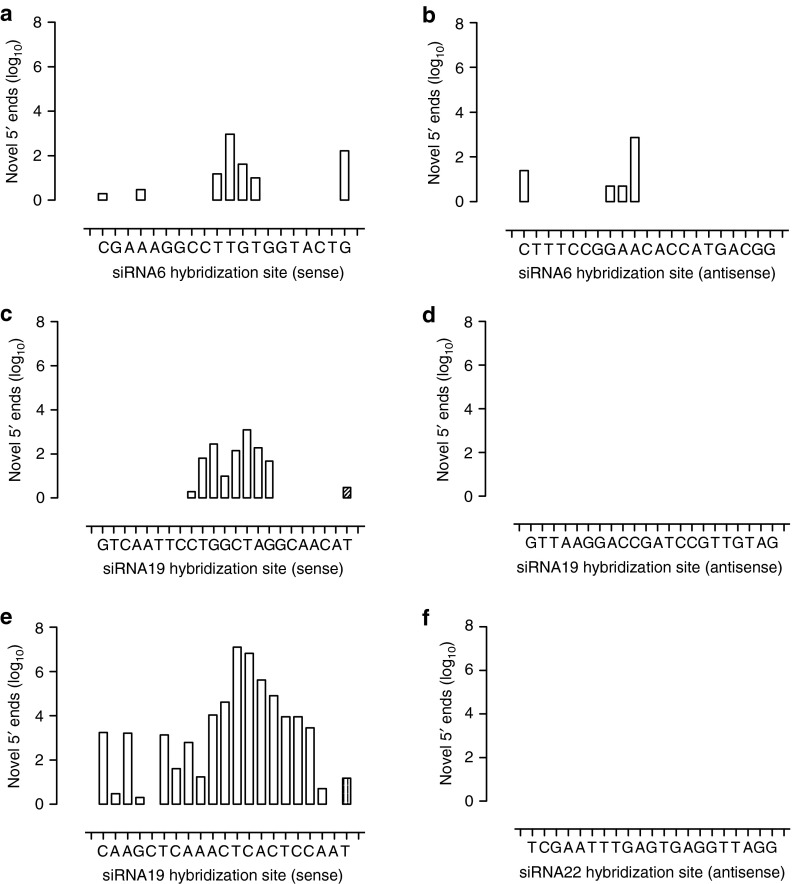
Figure 4.
Detection of single siRNA-mediated cleavage points in the sense and antisense strands of the HCV replicon RNA genome by RACE-Seq. Following in vitro transfection of HCV replicon cells with individual synthetic siRNAs, RACE-Seq was performed using gene-specific reverse transcription primers hybridizing immediately downstream of the target sites of (a,b) siRNA6, (c,d) siRNA19, or (e,f) siRNA22 on either (a,c,e) the sense or (b,d,f) the antisense RNA genomes of the HCV replicon. The incidence of novel 5′ ends (y axis) on the HCV replicon genome within each siRNA hybridization site in a given orientation (x axis) is described. The orientation of potential 5′→3′ RNA degradation is indicated using a right or left facing triangle.