Figure 2.
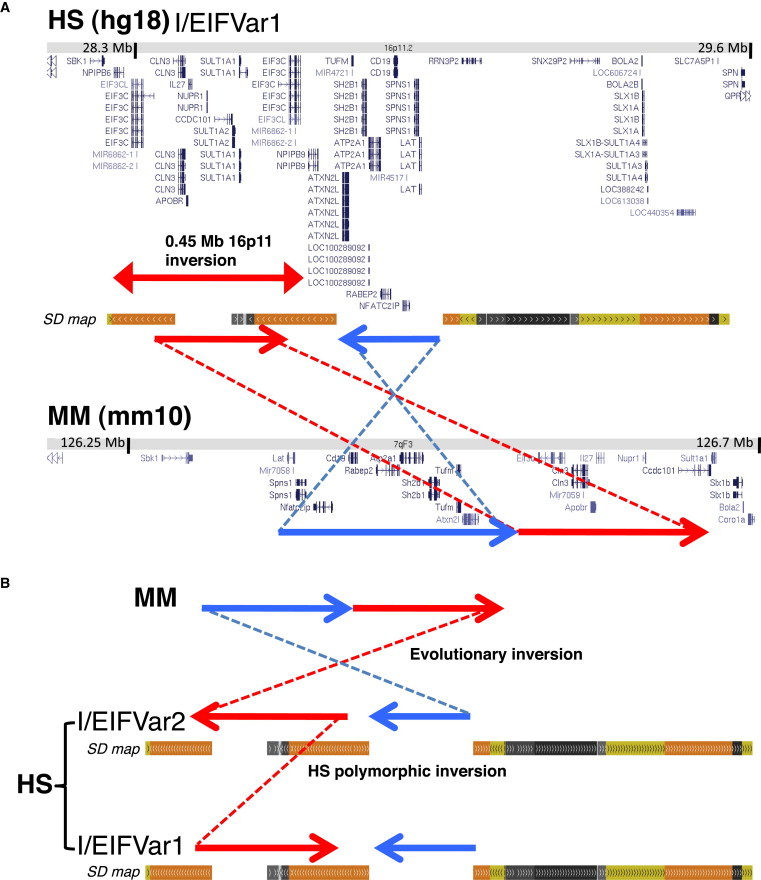
Comparative Mapping, Gene Content, and Predicted Evolution of the Inversion Polymorphism at 16p11.2
(A) Comparative mapping of the human (HS) reference region with the orthologous region in mouse (MM) showing the gene content and orientation according to the UCSC Genome Browser. The blue and red arrows with the connected lines illustrate the length and relative orientation of the blocks of conserved synteny between both species. Genomic structure in other sequenced mammals, such as cat, dog, and rat, is identical to that in mouse. Large and complex blocks of segmental duplications, absent in mouse, are observed in the human map at the breaks of synteny.
(B) Schematic representation of the predicted evolutionary inversion between the mouse genome (MM) and the likely most ancestral human allele (EIFVar2/I). The subsequent inversion between blocks of segmental duplications to generate the other allele (EIFVar1/NI) in humans (HS) is shown at the bottom.
